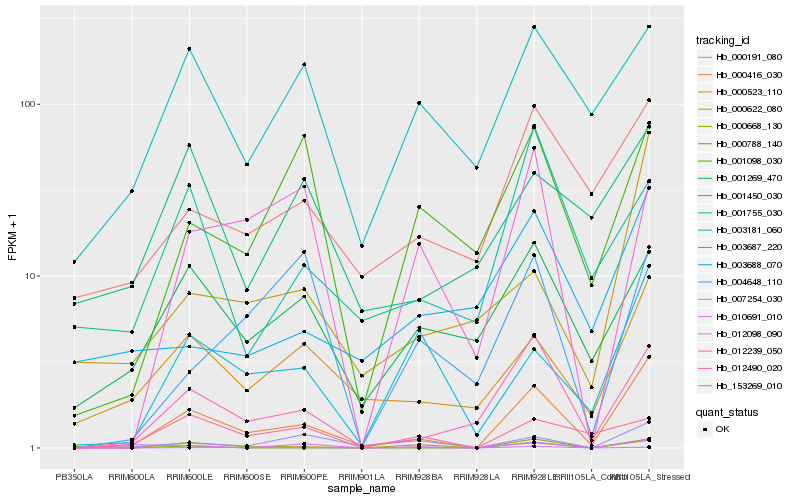
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000416_030 |
0.0 |
transcription factor |
TF Family: OFP |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_012490_020 |
0.2091225736 |
- |
- |
PREDICTED: protein phosphatase 2C 77 [Jatropha curcas] |
| 3 |
Hb_000523_110 |
0.255180563 |
- |
- |
PREDICTED: probable polyamine oxidase 4 [Jatropha curcas] |
| 4 |
Hb_007254_030 |
0.2596658545 |
- |
- |
kinase, putative [Ricinus communis] |
| 5 |
Hb_003687_220 |
0.2654614442 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0008s16660g [Populus trichocarpa] |
| 6 |
Hb_000622_080 |
0.2836754181 |
- |
- |
auxin-responsive GH3 family protein [Hevea brasiliensis] |
| 7 |
Hb_000191_080 |
0.2903443412 |
- |
- |
PREDICTED: uncharacterized protein LOC105637945 [Jatropha curcas] |
| 8 |
Hb_000668_130 |
0.2951284106 |
- |
- |
- |
| 9 |
Hb_010691_010 |
0.2955537511 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 10 |
Hb_001269_470 |
0.3069815243 |
- |
- |
PREDICTED: uncharacterized protein LOC105630313 [Jatropha curcas] |
| 11 |
Hb_001098_030 |
0.3100866871 |
- |
- |
PREDICTED: probable NAD(P)H dehydrogenase (quinone) FQR1-like 1 [Populus euphratica] |
| 12 |
Hb_003181_060 |
0.3123900856 |
- |
- |
PREDICTED: triosephosphate isomerase, cytosolic [Jatropha curcas] |
| 13 |
Hb_012098_090 |
0.3161795801 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 14 |
Hb_000788_140 |
0.3171849275 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_003688_070 |
0.3189457453 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein E [Jatropha curcas] |
| 16 |
Hb_153269_010 |
0.3191214201 |
- |
- |
hypothetical protein CICLE_v10029937mg [Citrus clementina] |
| 17 |
Hb_012239_050 |
0.3193248613 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001755_030 |
0.3267977175 |
- |
- |
PREDICTED: D-glycerate 3-kinase, chloroplastic-like [Populus euphratica] |
| 19 |
Hb_001450_030 |
0.3273505658 |
- |
- |
PREDICTED: uncharacterized protein LOC105650827 [Jatropha curcas] |
| 20 |
Hb_004648_110 |
0.3300399213 |
- |
- |
ATP binding protein, putative [Ricinus communis] |