| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
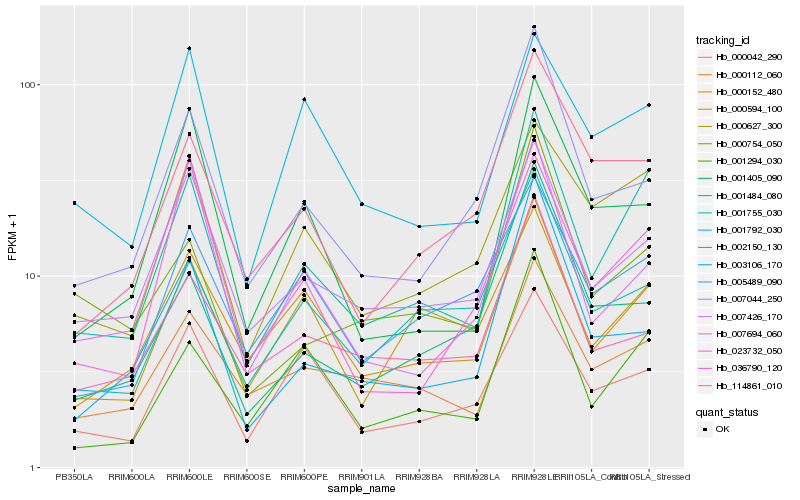
Hb_001755_030 |
0.0 |
- |
- |
PREDICTED: D-glycerate 3-kinase, chloroplastic-like [Populus euphratica] |
| 2 |
Hb_000627_300 |
0.1353020255 |
- |
- |
PREDICTED: uncharacterized protein LOC105632672 [Jatropha curcas] |
| 3 |
Hb_001294_030 |
0.1360247055 |
- |
- |
PREDICTED: uncharacterized protein LOC105644307 [Jatropha curcas] |
| 4 |
Hb_000152_480 |
0.1435448714 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 2, chloroplastic [Jatropha curcas] |
| 5 |
Hb_007426_170 |
0.1507890029 |
transcription factor |
TF Family: TCP |
- |
| 6 |
Hb_000112_060 |
0.154726051 |
- |
- |
PREDICTED: thioredoxin-like protein CITRX, chloroplastic [Jatropha curcas] |
| 7 |
Hb_114861_010 |
0.1627493211 |
- |
- |
STRESS ENHANCED protein 1 [Populus trichocarpa] |
| 8 |
Hb_005489_090 |
0.168767596 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase ICT1, mitochondrial [Jatropha curcas] |
| 9 |
Hb_001792_030 |
0.1699110162 |
- |
- |
PREDICTED: aminoacyl tRNA synthase complex-interacting multifunctional protein 1 [Jatropha curcas] |
| 10 |
Hb_007044_250 |
0.1707934882 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000754_050 |
0.1713799372 |
- |
- |
PREDICTED: thylakoid lumenal 19 kDa protein, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000594_100 |
0.171476911 |
- |
- |
PREDICTED: methionine aminopeptidase 1B, chloroplastic [Jatropha curcas] |
| 13 |
Hb_002150_130 |
0.1768253366 |
- |
- |
50S ribosomal protein L18, chloroplast precursor, putative [Ricinus communis] |
| 14 |
Hb_023732_050 |
0.1773330948 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 15 |
Hb_000042_290 |
0.1781180737 |
- |
- |
PREDICTED: uncharacterized protein LOC105632818 isoform X2 [Jatropha curcas] |
| 16 |
Hb_001405_090 |
0.1805680378 |
- |
- |
PREDICTED: photosynthetic NDH subunit of subcomplex B 5, chloroplastic [Jatropha curcas] |
| 17 |
Hb_007694_060 |
0.1820002235 |
- |
- |
PREDICTED: 50S ribosomal protein L35, chloroplastic [Jatropha curcas] |
| 18 |
Hb_036790_120 |
0.1821317537 |
- |
- |
PREDICTED: uncharacterized protein LOC105645122 isoform X1 [Jatropha curcas] |
| 19 |
Hb_003106_170 |
0.1827650983 |
- |
- |
Endonuclease III, putative [Ricinus communis] |
| 20 |
Hb_001484_080 |
0.1832719967 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |