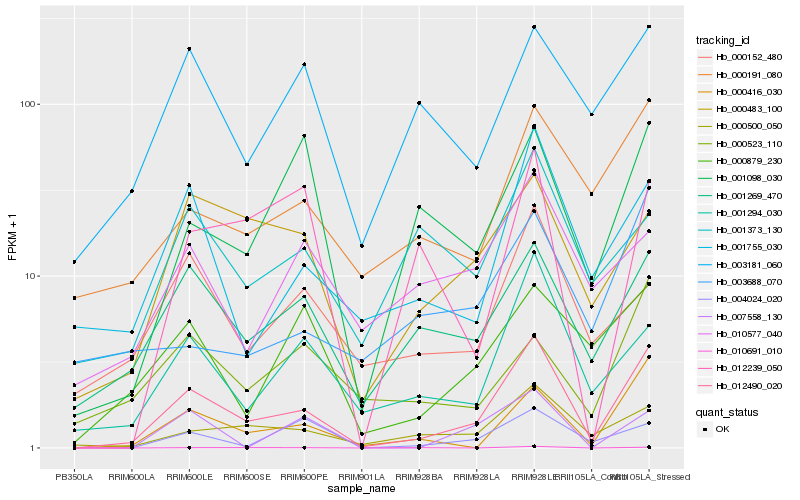
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012490_020 |
0.0 |
- |
- |
PREDICTED: protein phosphatase 2C 77 [Jatropha curcas] |
| 2 |
Hb_000416_030 |
0.2091225736 |
transcription factor |
TF Family: OFP |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000500_050 |
0.2210406785 |
- |
- |
PREDICTED: uncharacterized protein LOC105160092 [Sesamum indicum] |
| 4 |
Hb_001294_030 |
0.2309572467 |
- |
- |
PREDICTED: uncharacterized protein LOC105644307 [Jatropha curcas] |
| 5 |
Hb_001755_030 |
0.2374522808 |
- |
- |
PREDICTED: D-glycerate 3-kinase, chloroplastic-like [Populus euphratica] |
| 6 |
Hb_001269_470 |
0.2376748574 |
- |
- |
PREDICTED: uncharacterized protein LOC105630313 [Jatropha curcas] |
| 7 |
Hb_000191_080 |
0.2527364978 |
- |
- |
PREDICTED: uncharacterized protein LOC105637945 [Jatropha curcas] |
| 8 |
Hb_004024_020 |
0.2602760361 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 9 |
Hb_000879_230 |
0.2631573593 |
- |
- |
PREDICTED: tetratricopeptide repeat protein 7A [Jatropha curcas] |
| 10 |
Hb_010691_010 |
0.2644913765 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 11 |
Hb_000483_100 |
0.2648466473 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 9-like [Jatropha curcas] |
| 12 |
Hb_001098_030 |
0.2677642172 |
- |
- |
PREDICTED: probable NAD(P)H dehydrogenase (quinone) FQR1-like 1 [Populus euphratica] |
| 13 |
Hb_003181_060 |
0.2738345162 |
- |
- |
PREDICTED: triosephosphate isomerase, cytosolic [Jatropha curcas] |
| 14 |
Hb_000152_480 |
0.275129727 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 2, chloroplastic [Jatropha curcas] |
| 15 |
Hb_007558_130 |
0.277200307 |
- |
- |
- |
| 16 |
Hb_012239_050 |
0.2795140458 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003688_070 |
0.2839648366 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein E [Jatropha curcas] |
| 18 |
Hb_001373_130 |
0.2868694769 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000523_110 |
0.2880301644 |
- |
- |
PREDICTED: probable polyamine oxidase 4 [Jatropha curcas] |
| 20 |
Hb_010577_040 |
0.2884956636 |
- |
- |
PREDICTED: uncharacterized protein LOC105643738 [Jatropha curcas] |