| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
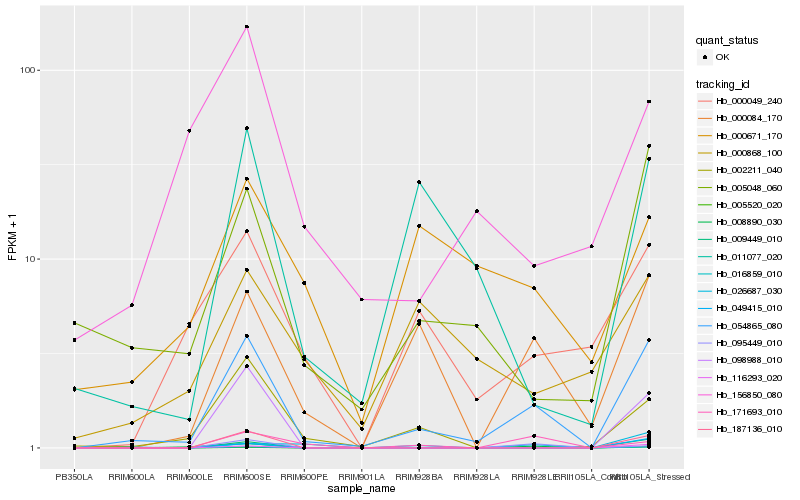
Hb_054865_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642789 [Jatropha curcas] |
| 2 |
Hb_000084_170 |
0.2678445223 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_005048_060 |
0.2923173087 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_008890_030 |
0.314303148 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 5 |
Hb_016859_010 |
0.3167744888 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 6 |
Hb_187136_010 |
0.3215158045 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 7 |
Hb_000049_240 |
0.3224778791 |
- |
- |
PREDICTED: uncharacterized protein LOC105644456 [Jatropha curcas] |
| 8 |
Hb_002211_040 |
0.3231873104 |
- |
- |
PREDICTED: subtilisin-like protease SBT3.5 [Jatropha curcas] |
| 9 |
Hb_009449_010 |
0.328412429 |
- |
- |
Serine/threonine-protein kinase HT1 [Gossypium arboreum] |
| 10 |
Hb_098988_010 |
0.3328041558 |
- |
- |
hypothetical protein JCGZ_18539 [Jatropha curcas] |
| 11 |
Hb_011077_020 |
0.3397904499 |
- |
- |
kinase, putative [Ricinus communis] |
| 12 |
Hb_116293_020 |
0.3423535011 |
- |
- |
Uncharacterized protein TCM_043787 [Theobroma cacao] |
| 13 |
Hb_005520_020 |
0.3427559965 |
- |
- |
hypothetical protein EUGRSUZ_G03336 [Eucalyptus grandis] |
| 14 |
Hb_026687_030 |
0.3489552601 |
- |
- |
hypothetical protein AMTR_s00038p00220080 [Amborella trichopoda] |
| 15 |
Hb_171693_010 |
0.3490301111 |
- |
- |
hypothetical protein JCGZ_26531 [Jatropha curcas] |
| 16 |
Hb_049415_010 |
0.3562568237 |
- |
- |
- |
| 17 |
Hb_156850_080 |
0.3620174339 |
- |
- |
PREDICTED: uncharacterized protein LOC105635247 [Jatropha curcas] |
| 18 |
Hb_095449_010 |
0.3645657198 |
- |
- |
PREDICTED: low-temperature-induced 65 kDa protein-like [Jatropha curcas] |
| 19 |
Hb_000868_100 |
0.3726300455 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RNF144A [Jatropha curcas] |
| 20 |
Hb_000671_170 |
0.3735307099 |
- |
- |
kinase, putative [Ricinus communis] |