| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
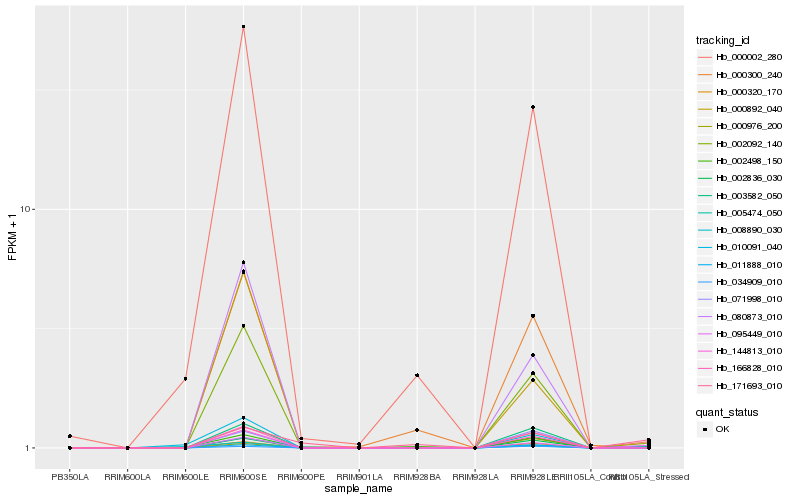
Hb_095449_010 |
0.0 |
- |
- |
PREDICTED: low-temperature-induced 65 kDa protein-like [Jatropha curcas] |
| 2 |
Hb_005474_050 |
0.2245879741 |
- |
- |
putative leucine-rich repeat transmembrane protein kinase [Populus trichocarpa] |
| 3 |
Hb_002498_150 |
0.2343808834 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002092_140 |
0.2660351561 |
- |
- |
hypothetical protein EUGRSUZ_B00708, partial [Eucalyptus grandis] |
| 5 |
Hb_166828_010 |
0.2661981246 |
- |
- |
- |
| 6 |
Hb_000320_170 |
0.2786664718 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_080873_010 |
0.2812142874 |
- |
- |
- |
| 8 |
Hb_000002_280 |
0.2824464894 |
- |
- |
PREDICTED: two-pore potassium channel 1 [Jatropha curcas] |
| 9 |
Hb_002836_030 |
0.2834805562 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 10 |
Hb_003582_050 |
0.2847563571 |
- |
- |
hypothetical protein POPTR_0008s13950g [Populus trichocarpa] |
| 11 |
Hb_144813_010 |
0.2895112934 |
- |
- |
PREDICTED: uncharacterized protein LOC105781565 [Gossypium raimondii] |
| 12 |
Hb_000300_240 |
0.2939706488 |
- |
- |
Sodium/potassium/calcium exchanger 6 precursor, putative [Ricinus communis] |
| 13 |
Hb_008890_030 |
0.2948083808 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 14 |
Hb_171693_010 |
0.2971590027 |
- |
- |
hypothetical protein JCGZ_26531 [Jatropha curcas] |
| 15 |
Hb_000892_040 |
0.3030525029 |
- |
- |
- |
| 16 |
Hb_011888_010 |
0.3032104178 |
- |
- |
Receptor like protein 46, putative [Theobroma cacao] |
| 17 |
Hb_071998_010 |
0.3050483023 |
- |
- |
PREDICTED: uncharacterized protein LOC105800957 [Gossypium raimondii] |
| 18 |
Hb_000976_200 |
0.3058720739 |
- |
- |
- |
| 19 |
Hb_010091_040 |
0.3161357879 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 20 |
Hb_034909_010 |
0.3190613227 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |