| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
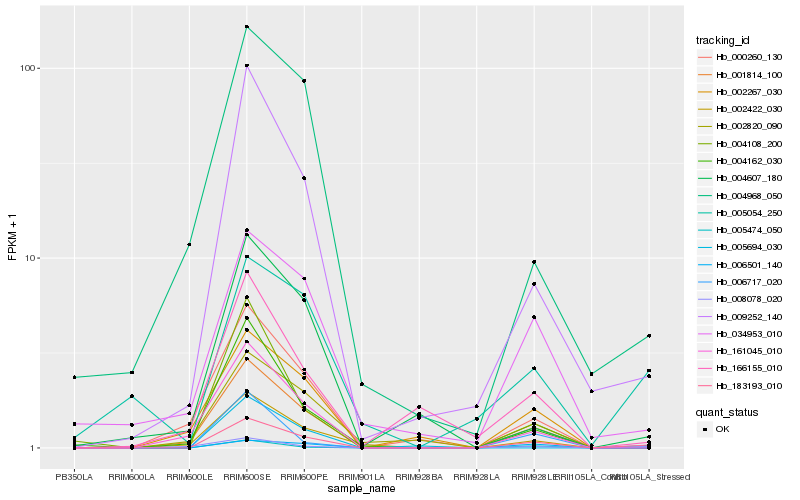
Hb_183193_010 |
0.0 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 2 |
Hb_009252_140 |
0.1850668419 |
- |
- |
Spotted leaf protein, putative [Ricinus communis] |
| 3 |
Hb_005474_050 |
0.1922159962 |
- |
- |
putative leucine-rich repeat transmembrane protein kinase [Populus trichocarpa] |
| 4 |
Hb_001814_100 |
0.1954283502 |
- |
- |
PREDICTED: uncharacterized protein LOC105119066 [Populus euphratica] |
| 5 |
Hb_161045_010 |
0.2303975323 |
- |
- |
hypothetical protein JCGZ_15100 [Jatropha curcas] |
| 6 |
Hb_006501_140 |
0.2329066787 |
- |
- |
PREDICTED: MATE efflux family protein 9-like [Jatropha curcas] |
| 7 |
Hb_004162_030 |
0.2395095225 |
- |
- |
PREDICTED: lysosomal Pro-X carboxypeptidase-like [Jatropha curcas] |
| 8 |
Hb_034953_010 |
0.242944252 |
- |
- |
hypothetical protein JCGZ_00031 [Jatropha curcas] |
| 9 |
Hb_002422_030 |
0.2435693836 |
- |
- |
E3 ubiquitin-protein ligase UPL5 -like protein [Gossypium arboreum] |
| 10 |
Hb_002267_030 |
0.2463241894 |
- |
- |
hypothetical protein JCGZ_24883 [Jatropha curcas] |
| 11 |
Hb_002820_090 |
0.2480559935 |
- |
- |
PREDICTED: WAT1-related protein At1g21890-like [Jatropha curcas] |
| 12 |
Hb_000260_130 |
0.2496182501 |
- |
- |
Acid phosphatase 1 precursor, putative [Ricinus communis] |
| 13 |
Hb_005054_250 |
0.251700179 |
- |
- |
PREDICTED: uncharacterized acetyltransferase At3g50280 [Jatropha curcas] |
| 14 |
Hb_004968_050 |
0.2533001834 |
- |
- |
PREDICTED: putative lipid-transfer protein DIR1 [Jatropha curcas] |
| 15 |
Hb_004108_200 |
0.2533939467 |
- |
- |
PREDICTED: adenylate isopentenyltransferase 3, chloroplastic [Jatropha curcas] |
| 16 |
Hb_166155_010 |
0.2585250525 |
- |
- |
PREDICTED: metalloendoproteinase 1 [Jatropha curcas] |
| 17 |
Hb_008078_020 |
0.2603352581 |
- |
- |
hypothetical protein JCGZ_01297 [Jatropha curcas] |
| 18 |
Hb_004607_180 |
0.2616201971 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_005694_030 |
0.2634605644 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase PUB22-like [Jatropha curcas] |
| 20 |
Hb_006717_020 |
0.2638565426 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |