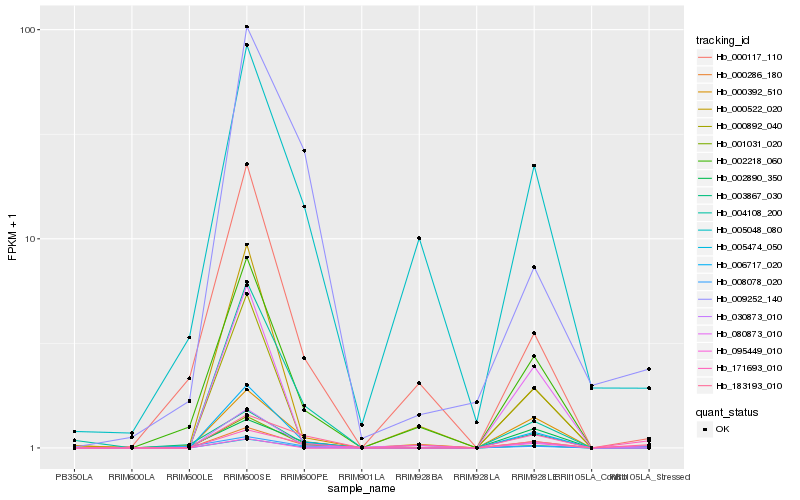
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005474_050 |
0.0 |
- |
- |
putative leucine-rich repeat transmembrane protein kinase [Populus trichocarpa] |
| 2 |
Hb_183193_010 |
0.1922159962 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 3 |
Hb_006717_020 |
0.2079072094 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 4 |
Hb_095449_010 |
0.2245879741 |
- |
- |
PREDICTED: low-temperature-induced 65 kDa protein-like [Jatropha curcas] |
| 5 |
Hb_008078_020 |
0.2529425432 |
- |
- |
hypothetical protein JCGZ_01297 [Jatropha curcas] |
| 6 |
Hb_003867_030 |
0.2575174733 |
- |
- |
- |
| 7 |
Hb_002218_060 |
0.2580421574 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002890_350 |
0.2586093974 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 9 |
Hb_000392_510 |
0.2704778152 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin of 36 kDa-like [Jatropha curcas] |
| 10 |
Hb_004108_200 |
0.276026248 |
- |
- |
PREDICTED: adenylate isopentenyltransferase 3, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000892_040 |
0.2762490445 |
- |
- |
- |
| 12 |
Hb_080873_010 |
0.2764863854 |
- |
- |
- |
| 13 |
Hb_000286_180 |
0.2796446562 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 4 [Jatropha curcas] |
| 14 |
Hb_171693_010 |
0.280108498 |
- |
- |
hypothetical protein JCGZ_26531 [Jatropha curcas] |
| 15 |
Hb_001031_020 |
0.2819070791 |
- |
- |
hypothetical protein VITISV_000631 [Vitis vinifera] |
| 16 |
Hb_005048_080 |
0.2826311474 |
- |
- |
PREDICTED: PAP-specific phosphatase HAL2-like [Jatropha curcas] |
| 17 |
Hb_000117_110 |
0.2827454009 |
- |
- |
transferase, putative [Ricinus communis] |
| 18 |
Hb_030873_010 |
0.2866549552 |
- |
- |
PREDICTED: putative cyclic nucleotide-gated ion channel 15 isoform X2 [Cucumis sativus] |
| 19 |
Hb_009252_140 |
0.2871010641 |
- |
- |
Spotted leaf protein, putative [Ricinus communis] |
| 20 |
Hb_000522_020 |
0.2879297285 |
- |
- |
PREDICTED: two-pore potassium channel 1 [Jatropha curcas] |