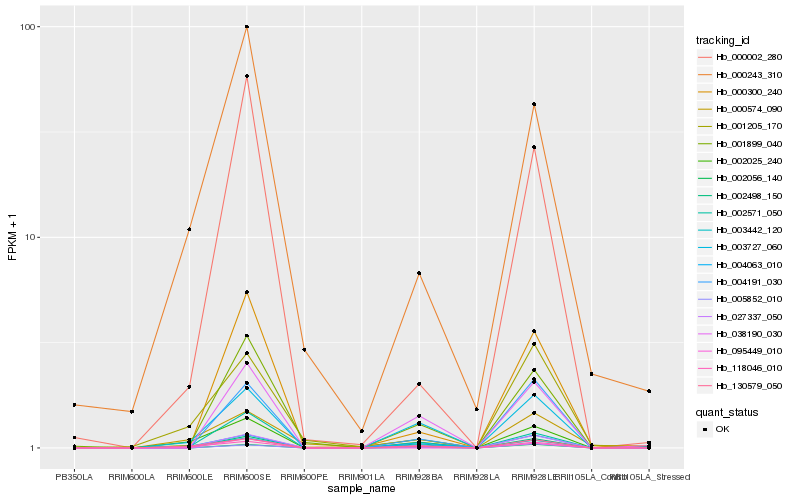
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002498_150 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_027337_050 |
0.2078032787 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 3 |
Hb_130579_050 |
0.214210252 |
- |
- |
hypothetical protein JCGZ_25577 [Jatropha curcas] |
| 4 |
Hb_002025_240 |
0.2174801034 |
- |
- |
- |
| 5 |
Hb_004191_030 |
0.2195057067 |
- |
- |
hypothetical protein POPTR_0013s13140g [Populus trichocarpa] |
| 6 |
Hb_001899_040 |
0.221979606 |
- |
- |
PREDICTED: cyclic nucleotide-gated ion channel 2-like [Jatropha curcas] |
| 7 |
Hb_002571_050 |
0.2260322851 |
- |
- |
PREDICTED: cation/calcium exchanger 1 [Jatropha curcas] |
| 8 |
Hb_003727_060 |
0.2266581238 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 9 |
Hb_002056_140 |
0.227856416 |
- |
- |
- |
| 10 |
Hb_000002_280 |
0.2286234308 |
- |
- |
PREDICTED: two-pore potassium channel 1 [Jatropha curcas] |
| 11 |
Hb_000300_240 |
0.2299565948 |
- |
- |
Sodium/potassium/calcium exchanger 6 precursor, putative [Ricinus communis] |
| 12 |
Hb_000243_310 |
0.2323814805 |
- |
- |
lactoylglutathione lyase, putative [Ricinus communis] |
| 13 |
Hb_095449_010 |
0.2343808834 |
- |
- |
PREDICTED: low-temperature-induced 65 kDa protein-like [Jatropha curcas] |
| 14 |
Hb_004063_010 |
0.2359421754 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 15 |
Hb_003442_120 |
0.2367846336 |
- |
- |
hypothetical protein VITISV_019818 [Vitis vinifera] |
| 16 |
Hb_005852_010 |
0.2376646771 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 17 |
Hb_118046_010 |
0.2411292251 |
- |
- |
PREDICTED: uncharacterized protein LOC102596706 [Solanum tuberosum] |
| 18 |
Hb_000574_090 |
0.2420601653 |
- |
- |
PREDICTED: uncharacterized protein LOC104878039 [Vitis vinifera] |
| 19 |
Hb_038190_030 |
0.244329792 |
- |
- |
- |
| 20 |
Hb_001205_170 |
0.2485774015 |
- |
- |
PREDICTED: uncharacterized protein LOC105633940 isoform X1 [Jatropha curcas] |