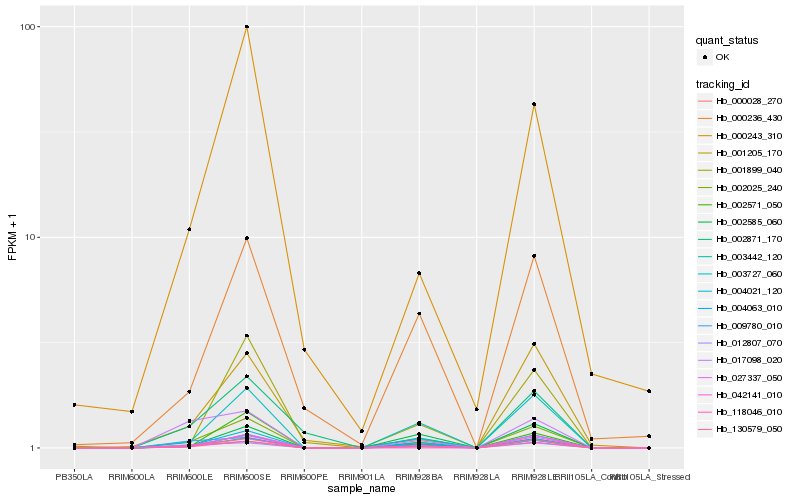
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002025_240 |
0.0 |
- |
- |
- |
| 2 |
Hb_130579_050 |
0.0459635519 |
- |
- |
hypothetical protein JCGZ_25577 [Jatropha curcas] |
| 3 |
Hb_003442_120 |
0.0755227106 |
- |
- |
hypothetical protein VITISV_019818 [Vitis vinifera] |
| 4 |
Hb_118046_010 |
0.0906856549 |
- |
- |
PREDICTED: uncharacterized protein LOC102596706 [Solanum tuberosum] |
| 5 |
Hb_004063_010 |
0.1340596358 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_003727_060 |
0.1402119412 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 7 |
Hb_027337_050 |
0.1627962681 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 8 |
Hb_012807_070 |
0.1671864346 |
- |
- |
PREDICTED: UDP-glycosyltransferase 86A2 [Jatropha curcas] |
| 9 |
Hb_000028_270 |
0.1706430001 |
- |
- |
- |
| 10 |
Hb_002571_050 |
0.1853257779 |
- |
- |
PREDICTED: cation/calcium exchanger 1 [Jatropha curcas] |
| 11 |
Hb_000243_310 |
0.1878887356 |
- |
- |
lactoylglutathione lyase, putative [Ricinus communis] |
| 12 |
Hb_009780_010 |
0.1903843429 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000236_430 |
0.1968826079 |
- |
- |
PREDICTED: uncharacterized protein LOC105633302 [Jatropha curcas] |
| 14 |
Hb_017098_020 |
0.199239218 |
- |
- |
Zinc finger protein GIS2 [Medicago truncatula] |
| 15 |
Hb_004021_120 |
0.2000107957 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 16 |
Hb_002871_170 |
0.2004949349 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein 21/22, putative [Ricinus communis] |
| 17 |
Hb_001899_040 |
0.2051975405 |
- |
- |
PREDICTED: cyclic nucleotide-gated ion channel 2-like [Jatropha curcas] |
| 18 |
Hb_001205_170 |
0.2067266518 |
- |
- |
PREDICTED: uncharacterized protein LOC105633940 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002585_060 |
0.2081412644 |
- |
- |
PREDICTED: uncharacterized protein LOC104899128, partial [Beta vulgaris subsp. vulgaris] |
| 20 |
Hb_042141_010 |
0.2113191255 |
- |
- |
- |