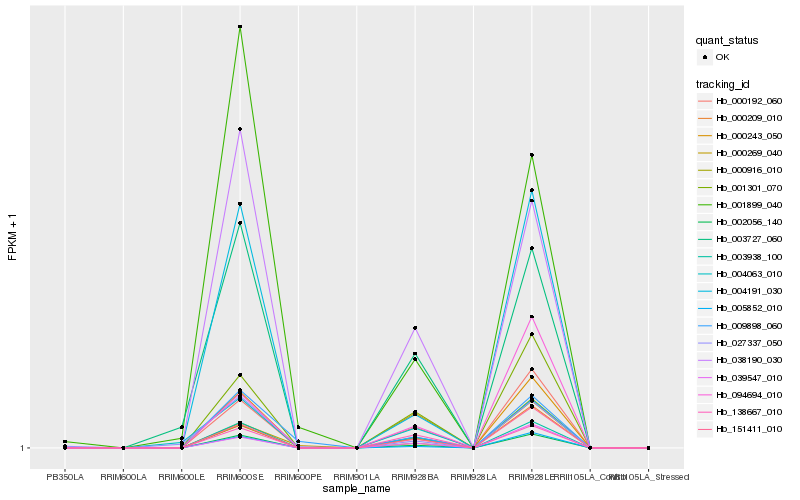
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002056_140 |
0.0 |
- |
- |
- |
| 2 |
Hb_004191_030 |
0.0756887389 |
- |
- |
hypothetical protein POPTR_0013s13140g [Populus trichocarpa] |
| 3 |
Hb_005852_010 |
0.079439082 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 4 |
Hb_003938_100 |
0.0999869197 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_151411_010 |
0.103179729 |
- |
- |
- |
| 6 |
Hb_000192_060 |
0.1060652487 |
- |
- |
- |
| 7 |
Hb_038190_030 |
0.1122303171 |
- |
- |
- |
| 8 |
Hb_001301_070 |
0.1145658909 |
- |
- |
- |
| 9 |
Hb_000209_010 |
0.1199301106 |
- |
- |
- |
| 10 |
Hb_000916_010 |
0.1448571343 |
- |
- |
hypothetical protein POPTR_0001s08730g [Populus trichocarpa] |
| 11 |
Hb_039547_010 |
0.1454480187 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 3-like [Jatropha curcas] |
| 12 |
Hb_000243_050 |
0.1460026192 |
- |
- |
ADP-ribosylation factor, putative [Ricinus communis] |
| 13 |
Hb_003727_060 |
0.1570554615 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 14 |
Hb_009898_060 |
0.1615441038 |
- |
- |
hypothetical protein POPTR_0007s09865g [Populus trichocarpa] |
| 15 |
Hb_138667_010 |
0.169990694 |
- |
- |
hypothetical protein JCGZ_04974 [Jatropha curcas] |
| 16 |
Hb_000269_040 |
0.1739985664 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 17 |
Hb_004063_010 |
0.1740235213 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 18 |
Hb_094694_010 |
0.175725459 |
- |
- |
- |
| 19 |
Hb_027337_050 |
0.1761067243 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 20 |
Hb_001899_040 |
0.1774533712 |
- |
- |
PREDICTED: cyclic nucleotide-gated ion channel 2-like [Jatropha curcas] |