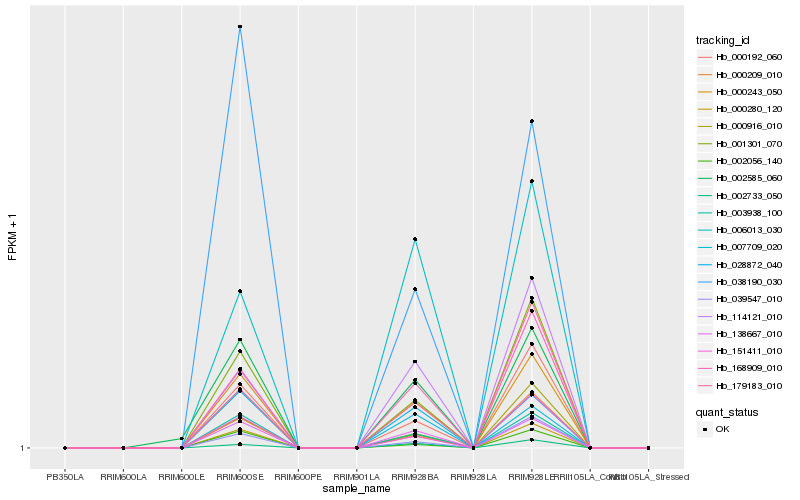
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000243_050 |
0.0 |
- |
- |
ADP-ribosylation factor, putative [Ricinus communis] |
| 2 |
Hb_138667_010 |
0.039797641 |
- |
- |
hypothetical protein JCGZ_04974 [Jatropha curcas] |
| 3 |
Hb_000280_120 |
0.0406849948 |
- |
- |
PREDICTED: uncharacterized protein LOC105643934 [Jatropha curcas] |
| 4 |
Hb_003938_100 |
0.0583950688 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_168909_010 |
0.0665428085 |
- |
- |
PREDICTED: uncharacterized protein LOC105640156 [Jatropha curcas] |
| 6 |
Hb_001301_070 |
0.0782128477 |
- |
- |
- |
| 7 |
Hb_028872_040 |
0.0882340502 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 8 |
Hb_000192_060 |
0.101067387 |
- |
- |
- |
| 9 |
Hb_007709_020 |
0.1162983439 |
- |
- |
PREDICTED: uncharacterized protein LOC105049010 [Elaeis guineensis] |
| 10 |
Hb_114121_010 |
0.1186847566 |
- |
- |
- |
| 11 |
Hb_151411_010 |
0.120548415 |
- |
- |
- |
| 12 |
Hb_002585_060 |
0.1218349898 |
- |
- |
PREDICTED: uncharacterized protein LOC104899128, partial [Beta vulgaris subsp. vulgaris] |
| 13 |
Hb_000209_010 |
0.1250701254 |
- |
- |
- |
| 14 |
Hb_002733_050 |
0.1256682638 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 15 |
Hb_006013_030 |
0.1342791598 |
- |
- |
- |
| 16 |
Hb_179183_010 |
0.1417414234 |
- |
- |
RALF-LIKE 32 family protein [Populus trichocarpa] |
| 17 |
Hb_000916_010 |
0.142898068 |
- |
- |
hypothetical protein POPTR_0001s08730g [Populus trichocarpa] |
| 18 |
Hb_038190_030 |
0.1431715826 |
- |
- |
- |
| 19 |
Hb_039547_010 |
0.1437826924 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 3-like [Jatropha curcas] |
| 20 |
Hb_002056_140 |
0.1460026192 |
- |
- |
- |