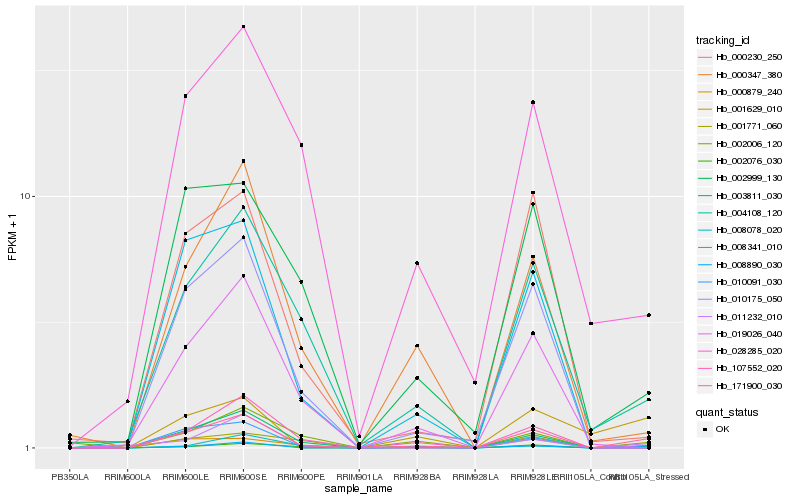
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_171900_030 |
0.0 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein MYBAS2 [Jatropha curcas] |
| 2 |
Hb_008890_030 |
0.1668962343 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 3 |
Hb_002076_030 |
0.1767847489 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 4 |
Hb_000879_240 |
0.2131300535 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105631008 [Jatropha curcas] |
| 5 |
Hb_004108_120 |
0.2164252543 |
- |
- |
PREDICTED: EID1-like F-box protein 3 [Jatropha curcas] |
| 6 |
Hb_011232_010 |
0.2353849843 |
- |
- |
Leucine-rich repeat receptor-like protein kinase family protein [Theobroma cacao] |
| 7 |
Hb_001771_060 |
0.2397969884 |
- |
- |
hypothetical protein B456_012G144100 [Gossypium raimondii] |
| 8 |
Hb_000347_380 |
0.2421106997 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_010175_050 |
0.253215722 |
- |
- |
PREDICTED: uncharacterized protein LOC105639906 [Jatropha curcas] |
| 10 |
Hb_008078_020 |
0.2549634534 |
- |
- |
hypothetical protein JCGZ_01297 [Jatropha curcas] |
| 11 |
Hb_019026_040 |
0.256361277 |
- |
- |
S-locus lectin protein kinase family protein [Theobroma cacao] |
| 12 |
Hb_002006_120 |
0.2573275058 |
- |
- |
PREDICTED: glycine--tRNA ligase 1, mitochondrial-like [Jatropha curcas] |
| 13 |
Hb_001629_010 |
0.2597198238 |
- |
- |
Disease resistance protein RPM1, putative [Theobroma cacao] |
| 14 |
Hb_002999_130 |
0.2597437027 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0008s08740g [Populus trichocarpa] |
| 15 |
Hb_003811_030 |
0.2612838226 |
- |
- |
hypothetical protein JCGZ_01307 [Jatropha curcas] |
| 16 |
Hb_028285_020 |
0.2614622152 |
transcription factor |
TF Family: C2C2-CO-like |
zinc finger protein, putative [Ricinus communis] |
| 17 |
Hb_008341_010 |
0.2630623964 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein HD1-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_000230_250 |
0.2646585898 |
- |
- |
PREDICTED: pyruvate, phosphate dikinase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_107552_020 |
0.2669919194 |
- |
- |
Ethylene-responsive transcription factor 1B, putative [Ricinus communis] |
| 20 |
Hb_010091_030 |
0.2678467153 |
- |
- |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |