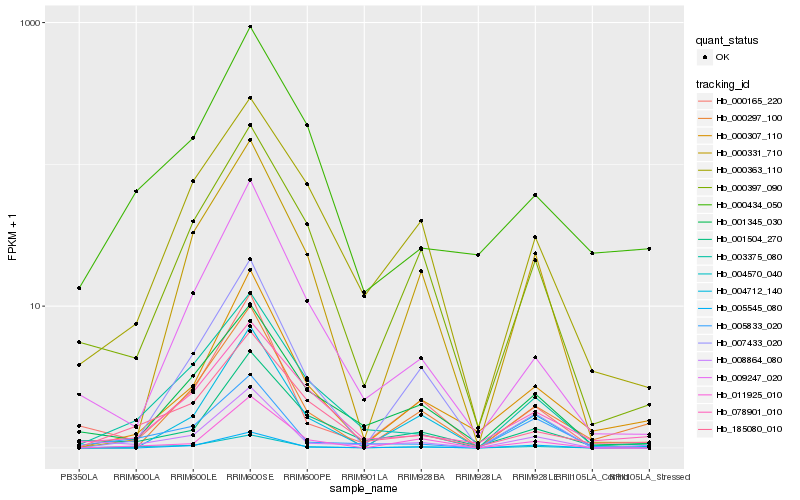
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004570_040 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_185080_010 |
0.1576003333 |
- |
- |
- |
| 3 |
Hb_003375_080 |
0.1778126532 |
- |
- |
PREDICTED: uncharacterized protein LOC105643382 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000307_110 |
0.1823262123 |
- |
- |
hypothetical protein JCGZ_08160 [Jatropha curcas] |
| 5 |
Hb_000397_090 |
0.1918846076 |
- |
- |
PREDICTED: uncharacterized protein LOC105641235 [Jatropha curcas] |
| 6 |
Hb_005545_080 |
0.1923097813 |
- |
- |
hypothetical protein JCGZ_01293 [Jatropha curcas] |
| 7 |
Hb_000434_050 |
0.1969060216 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 5 [Hevea brasiliensis] |
| 8 |
Hb_007433_020 |
0.2003987595 |
- |
- |
hypothetical protein VITISV_005320 [Vitis vinifera] |
| 9 |
Hb_000363_110 |
0.2027684556 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 21 [Jatropha curcas] |
| 10 |
Hb_011925_010 |
0.2034358453 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000297_100 |
0.2039622071 |
transcription factor |
TF Family: NAC |
NAC transcription factor 038 [Jatropha curcas] |
| 12 |
Hb_004712_140 |
0.2045094352 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 13 |
Hb_001345_030 |
0.2049385448 |
- |
- |
hypothetical protein POPTR_0007s10390g, partial [Populus trichocarpa] |
| 14 |
Hb_000165_220 |
0.2061230397 |
- |
- |
PREDICTED: taxadien-5-alpha-ol O-acetyltransferase [Jatropha curcas] |
| 15 |
Hb_078901_010 |
0.2071763814 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 16 |
Hb_001504_270 |
0.2076614332 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |
| 17 |
Hb_008864_080 |
0.2084533609 |
- |
- |
PREDICTED: uncharacterized protein LOC105631804 [Jatropha curcas] |
| 18 |
Hb_000331_710 |
0.210053098 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 19 |
Hb_009247_020 |
0.2107583431 |
- |
- |
hypothetical protein JCGZ_04985 [Jatropha curcas] |
| 20 |
Hb_005833_020 |
0.2109322598 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 [Jatropha curcas] |