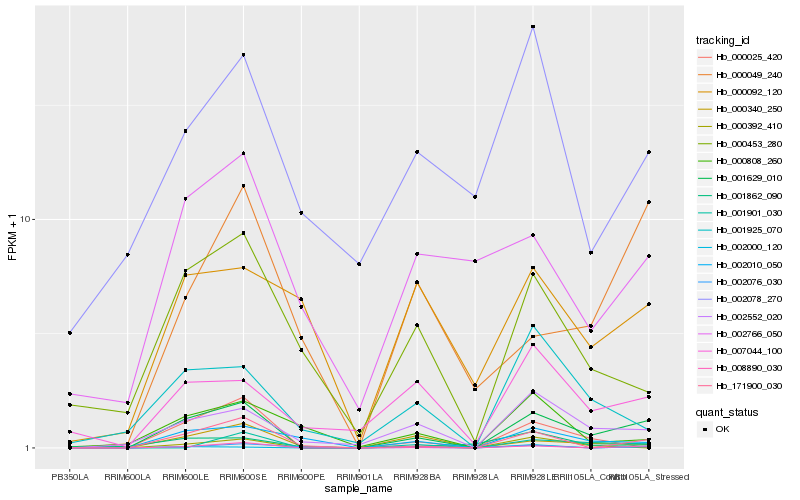
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001629_010 |
0.0 |
- |
- |
Disease resistance protein RPM1, putative [Theobroma cacao] |
| 2 |
Hb_008890_030 |
0.240729063 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 3 |
Hb_002552_020 |
0.2488179548 |
- |
- |
PREDICTED: 116 kDa U5 small nuclear ribonucleoprotein component-like [Jatropha curcas] |
| 4 |
Hb_171900_030 |
0.2597198238 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein MYBAS2 [Jatropha curcas] |
| 5 |
Hb_002010_050 |
0.2786848092 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000340_250 |
0.2849447387 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 4 [Jatropha curcas] |
| 7 |
Hb_007044_100 |
0.3055801338 |
- |
- |
hypothetical protein POPTR_0004s10420g [Populus trichocarpa] |
| 8 |
Hb_000025_420 |
0.3111245372 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001925_070 |
0.3145222249 |
- |
- |
PREDICTED: 40S ribosomal protein S3-3-like [Populus euphratica] |
| 10 |
Hb_002076_030 |
0.3175163401 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 11 |
Hb_000453_280 |
0.3193425491 |
- |
- |
negative cofactor 2 transcriptional co-repressor, putative [Ricinus communis] |
| 12 |
Hb_000049_240 |
0.3237721964 |
- |
- |
PREDICTED: uncharacterized protein LOC105644456 [Jatropha curcas] |
| 13 |
Hb_000392_410 |
0.324581536 |
- |
- |
PREDICTED: uncharacterized protein LOC104882159 [Vitis vinifera] |
| 14 |
Hb_002000_120 |
0.3287499012 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 15 |
Hb_002766_050 |
0.3324789501 |
- |
- |
PREDICTED: probable carboxylesterase 16 [Jatropha curcas] |
| 16 |
Hb_001901_030 |
0.3357944669 |
- |
- |
- |
| 17 |
Hb_002078_270 |
0.3362716021 |
- |
- |
PREDICTED: F-box protein At1g67340 [Jatropha curcas] |
| 18 |
Hb_001862_090 |
0.3368096364 |
- |
- |
hypothetical protein VITISV_026680 [Vitis vinifera] |
| 19 |
Hb_000808_260 |
0.3370525787 |
- |
- |
hypothetical protein RCOM_1434920 [Ricinus communis] |
| 20 |
Hb_000092_120 |
0.3382406954 |
- |
- |
PREDICTED: non-structural maintenance of chromosomes element 4 homolog A-like [Populus euphratica] |