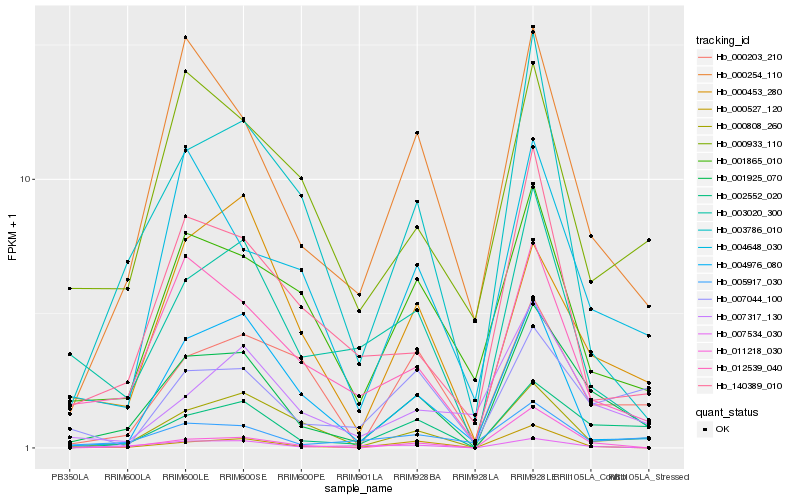
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001925_070 |
0.0 |
- |
- |
PREDICTED: 40S ribosomal protein S3-3-like [Populus euphratica] |
| 2 |
Hb_002552_020 |
0.1423958433 |
- |
- |
PREDICTED: 116 kDa U5 small nuclear ribonucleoprotein component-like [Jatropha curcas] |
| 3 |
Hb_000254_110 |
0.1864215531 |
- |
- |
PREDICTED: dynein light chain 1, cytoplasmic-like [Jatropha curcas] |
| 4 |
Hb_000808_260 |
0.1981337868 |
- |
- |
hypothetical protein RCOM_1434920 [Ricinus communis] |
| 5 |
Hb_012539_040 |
0.2029873871 |
transcription factor |
TF Family: CPP |
hypothetical protein JCGZ_07765 [Jatropha curcas] |
| 6 |
Hb_004648_030 |
0.2067887851 |
- |
- |
PREDICTED: probable dimethyladenosine transferase [Jatropha curcas] |
| 7 |
Hb_005917_030 |
0.2090487122 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000527_120 |
0.2127094383 |
- |
- |
hypothetical protein VITISV_027174 [Vitis vinifera] |
| 9 |
Hb_000453_280 |
0.2138729446 |
- |
- |
negative cofactor 2 transcriptional co-repressor, putative [Ricinus communis] |
| 10 |
Hb_001865_010 |
0.2178311357 |
transcription factor |
TF Family: B3 |
hypothetical protein RCOM_0465210 [Ricinus communis] |
| 11 |
Hb_007317_130 |
0.2217541563 |
- |
- |
retrotransposon protein, putative, unclassified [Oryza sativa Japonica Group] |
| 12 |
Hb_000203_210 |
0.2239728477 |
- |
- |
PREDICTED: uncharacterized protein LOC105040619 isoform X3 [Elaeis guineensis] |
| 13 |
Hb_003786_010 |
0.2241045645 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 2 isoform X2 [Jatropha curcas] |
| 14 |
Hb_140389_010 |
0.2247607056 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 15 |
Hb_003020_300 |
0.2255859313 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_007044_100 |
0.2260572081 |
- |
- |
hypothetical protein POPTR_0004s10420g [Populus trichocarpa] |
| 17 |
Hb_007534_030 |
0.2269282718 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 18 |
Hb_000933_110 |
0.2358680929 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 3 [Jatropha curcas] |
| 19 |
Hb_004976_080 |
0.2370137464 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 20 |
Hb_011218_030 |
0.2372159122 |
- |
- |
hypothetical protein VITISV_002891 [Vitis vinifera] |