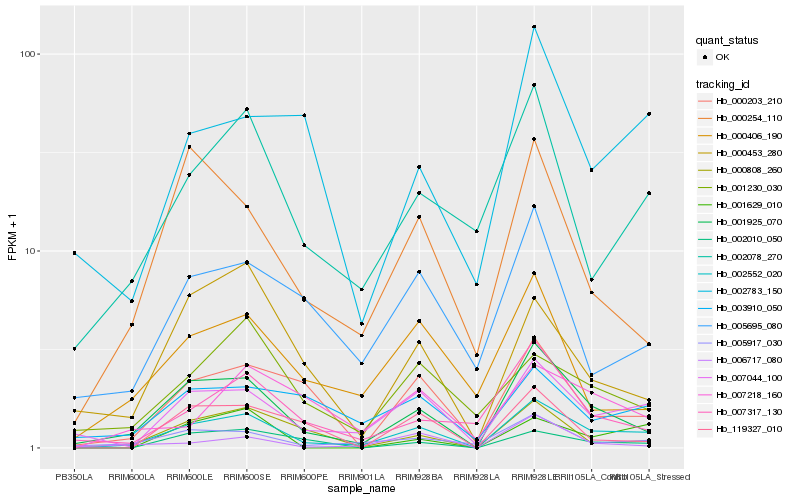
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002552_020 |
0.0 |
- |
- |
PREDICTED: 116 kDa U5 small nuclear ribonucleoprotein component-like [Jatropha curcas] |
| 2 |
Hb_001925_070 |
0.1423958433 |
- |
- |
PREDICTED: 40S ribosomal protein S3-3-like [Populus euphratica] |
| 3 |
Hb_007044_100 |
0.1757941602 |
- |
- |
hypothetical protein POPTR_0004s10420g [Populus trichocarpa] |
| 4 |
Hb_005917_030 |
0.2192116694 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000203_210 |
0.219410375 |
- |
- |
PREDICTED: uncharacterized protein LOC105040619 isoform X3 [Elaeis guineensis] |
| 6 |
Hb_002078_270 |
0.2260822783 |
- |
- |
PREDICTED: F-box protein At1g67340 [Jatropha curcas] |
| 7 |
Hb_000808_260 |
0.2260850586 |
- |
- |
hypothetical protein RCOM_1434920 [Ricinus communis] |
| 8 |
Hb_002010_050 |
0.2282593428 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_007317_130 |
0.2409609481 |
- |
- |
retrotransposon protein, putative, unclassified [Oryza sativa Japonica Group] |
| 10 |
Hb_000254_110 |
0.241417469 |
- |
- |
PREDICTED: dynein light chain 1, cytoplasmic-like [Jatropha curcas] |
| 11 |
Hb_003910_050 |
0.2442547636 |
- |
- |
PREDICTED: uncharacterized protein LOC105646926 [Jatropha curcas] |
| 12 |
Hb_000453_280 |
0.2442671845 |
- |
- |
negative cofactor 2 transcriptional co-repressor, putative [Ricinus communis] |
| 13 |
Hb_005695_080 |
0.2478502934 |
- |
- |
PREDICTED: acetyl-CoA carboxylase 1-like [Jatropha curcas] |
| 14 |
Hb_001629_010 |
0.2488179548 |
- |
- |
Disease resistance protein RPM1, putative [Theobroma cacao] |
| 15 |
Hb_006717_080 |
0.2494257469 |
- |
- |
PREDICTED: glutamic acid-rich protein-like [Jatropha curcas] |
| 16 |
Hb_119327_010 |
0.2498601185 |
- |
- |
hypothetical protein POPTR_0075s00200g, partial [Populus trichocarpa] |
| 17 |
Hb_000406_190 |
0.2499083722 |
desease resistance |
Gene Name: NB-ARC |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 18 |
Hb_002783_150 |
0.2513578339 |
- |
- |
cyclophilin [Hevea brasiliensis] |
| 19 |
Hb_007218_160 |
0.2514005577 |
- |
- |
PREDICTED: uncharacterized protein LOC105108358 [Populus euphratica] |
| 20 |
Hb_001230_030 |
0.2524721313 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g07650 isoform X2 [Populus euphratica] |