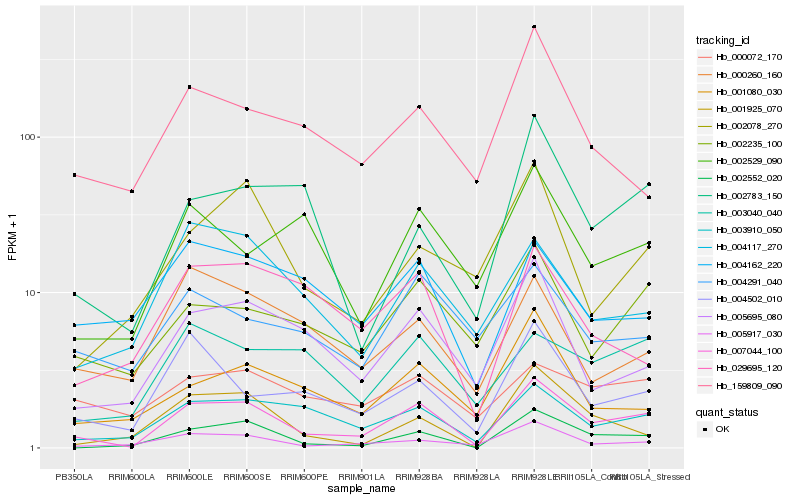
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007044_100 |
0.0 |
- |
- |
hypothetical protein POPTR_0004s10420g [Populus trichocarpa] |
| 2 |
Hb_002552_020 |
0.1757941602 |
- |
- |
PREDICTED: 116 kDa U5 small nuclear ribonucleoprotein component-like [Jatropha curcas] |
| 3 |
Hb_003910_050 |
0.1834188732 |
- |
- |
PREDICTED: uncharacterized protein LOC105646926 [Jatropha curcas] |
| 4 |
Hb_005917_030 |
0.1964365656 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_005695_080 |
0.2027981805 |
- |
- |
PREDICTED: acetyl-CoA carboxylase 1-like [Jatropha curcas] |
| 6 |
Hb_002235_100 |
0.2072627233 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 4 isoform X1 [Jatropha curcas] |
| 7 |
Hb_004502_010 |
0.2076904255 |
- |
- |
hypothetical protein JCGZ_23576 [Jatropha curcas] |
| 8 |
Hb_002783_150 |
0.217141412 |
- |
- |
cyclophilin [Hevea brasiliensis] |
| 9 |
Hb_001080_030 |
0.2208068806 |
- |
- |
AMP dependent ligase, putative [Ricinus communis] |
| 10 |
Hb_003040_040 |
0.2232351721 |
- |
- |
PREDICTED: protein SAWADEE HOMEODOMAIN HOMOLOG 1-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_004117_270 |
0.2253875388 |
- |
- |
- |
| 12 |
Hb_001925_070 |
0.2260572081 |
- |
- |
PREDICTED: 40S ribosomal protein S3-3-like [Populus euphratica] |
| 13 |
Hb_002529_090 |
0.2270019284 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase |
2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase [Hevea brasiliensis] |
| 14 |
Hb_002078_270 |
0.2291069423 |
- |
- |
PREDICTED: F-box protein At1g67340 [Jatropha curcas] |
| 15 |
Hb_029695_120 |
0.2304937101 |
- |
- |
hypothetical protein POPTR_0002s05320g [Populus trichocarpa] |
| 16 |
Hb_000072_170 |
0.2309186275 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 17 |
Hb_159809_090 |
0.2314727057 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-enyl diphosphate reductase |
4-hydroxy-3-methylbut-2-enyl diphosphate reductase [Hevea brasiliensis] |
| 18 |
Hb_004162_220 |
0.2319923238 |
- |
- |
PREDICTED: pheophorbide a oxygenase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_004291_040 |
0.23221181 |
- |
- |
hexokinase [Manihot esculenta] |
| 20 |
Hb_000260_160 |
0.2323609184 |
transcription factor |
TF Family: GNAT |
ATP binding protein, putative [Ricinus communis] |