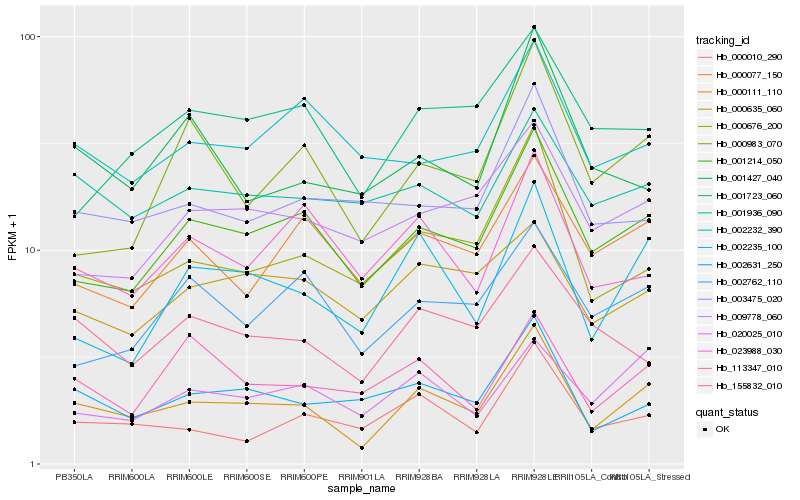
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000111_110 |
0.0 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 2 |
Hb_001214_050 |
0.1235618471 |
- |
- |
PREDICTED: bifunctional monothiol glutaredoxin-S16, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000676_200 |
0.1331615637 |
- |
- |
PREDICTED: CDK5RAP1-like protein [Jatropha curcas] |
| 4 |
Hb_002235_100 |
0.1361074145 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 4 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002631_250 |
0.14485706 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g20730 [Jatropha curcas] |
| 6 |
Hb_000010_290 |
0.1518748419 |
- |
- |
- |
| 7 |
Hb_020025_010 |
0.1543866474 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 homolog [Jatropha curcas] |
| 8 |
Hb_000077_150 |
0.158724573 |
- |
- |
PREDICTED: riboflavin synthase [Jatropha curcas] |
| 9 |
Hb_155832_010 |
0.159657785 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 10 |
Hb_001936_090 |
0.1616917342 |
- |
- |
PREDICTED: BOI-related E3 ubiquitin-protein ligase 1-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_000983_070 |
0.1622013085 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_023988_030 |
0.1632685744 |
- |
- |
PREDICTED: transmembrane protein 19 [Vitis vinifera] |
| 13 |
Hb_113347_010 |
0.1636060594 |
- |
- |
hypothetical protein CISIN_1g009336mg [Citrus sinensis] |
| 14 |
Hb_000635_060 |
0.1637161246 |
- |
- |
PREDICTED: uncharacterized protein LOC105644797 [Jatropha curcas] |
| 15 |
Hb_009778_060 |
0.1641424551 |
- |
- |
PREDICTED: uncharacterized protein LOC105636987 [Jatropha curcas] |
| 16 |
Hb_001723_060 |
0.1657156405 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
PREDICTED: LOW QUALITY PROTEIN: dihydrolipoyl dehydrogenase 2, mitochondrial [Populus euphratica] |
| 17 |
Hb_003475_020 |
0.1659094181 |
- |
- |
hypothetical protein JCGZ_13177 [Jatropha curcas] |
| 18 |
Hb_002762_110 |
0.1675851893 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001427_040 |
0.1679543034 |
- |
- |
PREDICTED: uncharacterized protein LOC105637784 [Jatropha curcas] |
| 20 |
Hb_002232_390 |
0.168556066 |
- |
- |
PREDICTED: uncharacterized protein LOC105635994 [Jatropha curcas] |