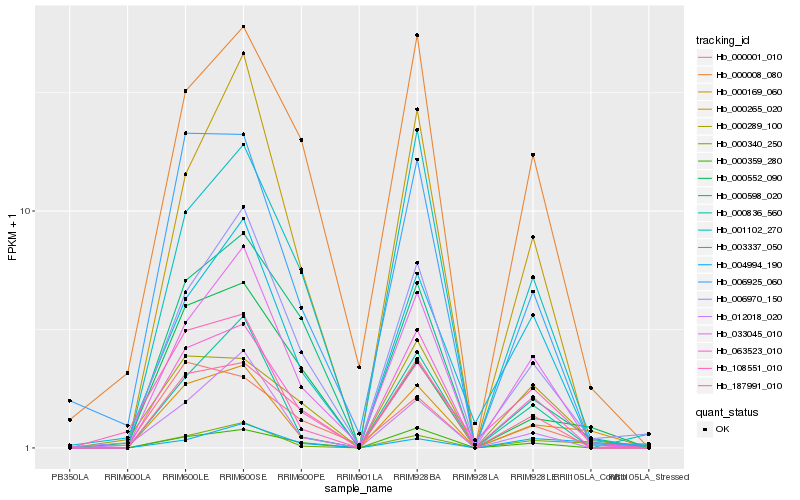
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000169_060 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_063523_010 |
0.1805506622 |
- |
- |
- |
| 3 |
Hb_033045_010 |
0.1857860543 |
- |
- |
copine, putative [Ricinus communis] |
| 4 |
Hb_000340_250 |
0.1930526936 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 4 [Jatropha curcas] |
| 5 |
Hb_108551_010 |
0.1942352118 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_006925_060 |
0.1957816559 |
- |
- |
hypothetical protein JCGZ_20651 [Jatropha curcas] |
| 7 |
Hb_006970_150 |
0.1971871383 |
- |
- |
PREDICTED: uncharacterized protein LOC105650471 [Jatropha curcas] |
| 8 |
Hb_004994_190 |
0.2100210034 |
- |
- |
PREDICTED: (+)-neomenthol dehydrogenase-like [Jatropha curcas] |
| 9 |
Hb_187991_010 |
0.2121386635 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 10 |
Hb_000265_020 |
0.2122500198 |
- |
- |
hypothetical protein POPTR_0011s16980g, partial [Populus trichocarpa] |
| 11 |
Hb_000552_090 |
0.2127342385 |
- |
- |
hypothetical protein JCGZ_18511 [Jatropha curcas] |
| 12 |
Hb_000359_280 |
0.2147765683 |
- |
- |
hypothetical protein OsI_15300 [Oryza sativa Indica Group] |
| 13 |
Hb_000289_100 |
0.2169334035 |
- |
- |
carbonyl reductase, putative [Ricinus communis] |
| 14 |
Hb_000001_010 |
0.2173914914 |
- |
- |
PREDICTED: uncharacterized protein LOC105648961 [Jatropha curcas] |
| 15 |
Hb_000008_080 |
0.2178889947 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_012018_020 |
0.2179301694 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK15 [Eucalyptus grandis] |
| 17 |
Hb_001102_270 |
0.2185516927 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Jatropha curcas] |
| 18 |
Hb_000836_560 |
0.2202901107 |
- |
- |
- |
| 19 |
Hb_000598_020 |
0.2206022607 |
- |
- |
PREDICTED: PRA1 family protein F2-like [Jatropha curcas] |
| 20 |
Hb_003337_050 |
0.2214367029 |
- |
- |
PREDICTED: probable protein phosphatase 2C 78 [Jatropha curcas] |