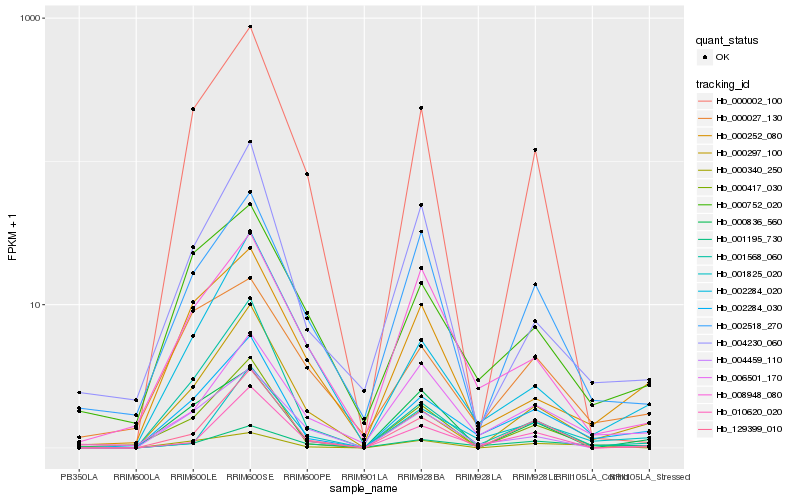
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002284_030 |
0.0 |
- |
- |
PREDICTED: xylem cysteine proteinase 2 [Jatropha curcas] |
| 2 |
Hb_006501_170 |
0.1888581218 |
- |
- |
hypothetical protein JCGZ_07371 [Jatropha curcas] |
| 3 |
Hb_000297_100 |
0.1916988862 |
transcription factor |
TF Family: NAC |
NAC transcription factor 038 [Jatropha curcas] |
| 4 |
Hb_000752_020 |
0.193361081 |
- |
- |
PREDICTED: uncharacterized protein LOC105641803 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000340_250 |
0.1960835646 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 4 [Jatropha curcas] |
| 6 |
Hb_000252_080 |
0.1980554756 |
- |
- |
PREDICTED: uncharacterized protein LOC105644644 [Jatropha curcas] |
| 7 |
Hb_004459_110 |
0.1988303237 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 isoform X2 [Populus euphratica] |
| 8 |
Hb_000417_030 |
0.199989376 |
transcription factor |
TF Family: G2-like |
PREDICTED: putative two-component response regulator ARR19 [Jatropha curcas] |
| 9 |
Hb_000836_560 |
0.2017346612 |
- |
- |
- |
| 10 |
Hb_002284_020 |
0.2018174711 |
- |
- |
PREDICTED: probable inorganic phosphate transporter 1-9 [Jatropha curcas] |
| 11 |
Hb_001568_060 |
0.2043892327 |
- |
- |
PREDICTED: uncharacterized protein LOC105650728 [Jatropha curcas] |
| 12 |
Hb_002518_270 |
0.2063849887 |
- |
- |
glycogen phosphorylase, putative [Ricinus communis] |
| 13 |
Hb_001195_730 |
0.2065455418 |
- |
- |
PREDICTED: protein LYK5-like [Jatropha curcas] |
| 14 |
Hb_129399_010 |
0.2076232241 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 15 |
Hb_000002_100 |
0.2096641696 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 16 |
Hb_001825_020 |
0.2103000319 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 17 |
Hb_004230_060 |
0.21089017 |
- |
- |
PREDICTED: ferritin, chloroplastic-like [Populus euphratica] |
| 18 |
Hb_010620_020 |
0.2109940431 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 19 |
Hb_008948_080 |
0.2117181534 |
- |
- |
PREDICTED: TBC1 domain family member 13-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_000027_130 |
0.2138184168 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 [Jatropha curcas] |