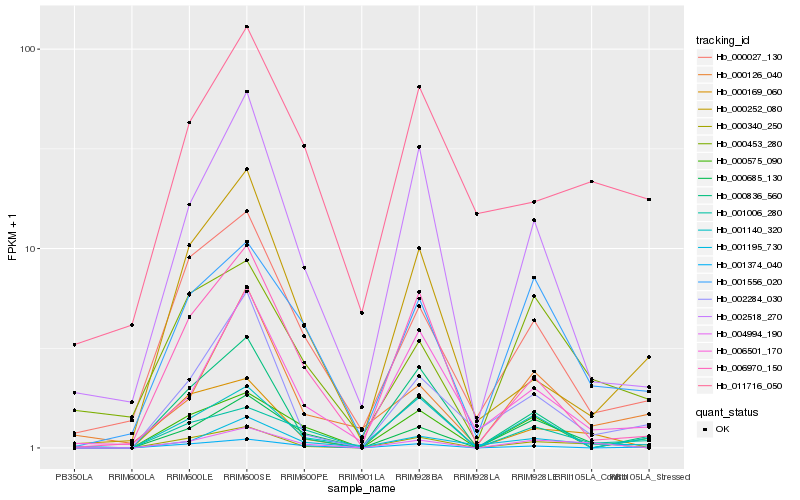
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000340_250 |
0.0 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 4 [Jatropha curcas] |
| 2 |
Hb_000169_060 |
0.1930526936 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002284_030 |
0.1960835646 |
- |
- |
PREDICTED: xylem cysteine proteinase 2 [Jatropha curcas] |
| 4 |
Hb_000836_560 |
0.2060514475 |
- |
- |
- |
| 5 |
Hb_004994_190 |
0.2063401342 |
- |
- |
PREDICTED: (+)-neomenthol dehydrogenase-like [Jatropha curcas] |
| 6 |
Hb_001556_020 |
0.2156147919 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 7 |
Hb_001140_320 |
0.2235467076 |
- |
- |
PREDICTED: oxalate--CoA ligase [Jatropha curcas] |
| 8 |
Hb_001374_040 |
0.2263163085 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Gossypium raimondii] |
| 9 |
Hb_002518_270 |
0.2274949995 |
- |
- |
glycogen phosphorylase, putative [Ricinus communis] |
| 10 |
Hb_000252_080 |
0.2319009987 |
- |
- |
PREDICTED: uncharacterized protein LOC105644644 [Jatropha curcas] |
| 11 |
Hb_000027_130 |
0.2320656767 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 [Jatropha curcas] |
| 12 |
Hb_006501_170 |
0.2355785487 |
- |
- |
hypothetical protein JCGZ_07371 [Jatropha curcas] |
| 13 |
Hb_001006_280 |
0.2365599401 |
- |
- |
PREDICTED: glutamate decarboxylase 4 [Jatropha curcas] |
| 14 |
Hb_006970_150 |
0.2381033204 |
- |
- |
PREDICTED: uncharacterized protein LOC105650471 [Jatropha curcas] |
| 15 |
Hb_000575_090 |
0.2397749761 |
- |
- |
PREDICTED: probable ubiquitin-conjugating enzyme E2 24 [Populus euphratica] |
| 16 |
Hb_000453_280 |
0.2403329228 |
- |
- |
negative cofactor 2 transcriptional co-repressor, putative [Ricinus communis] |
| 17 |
Hb_001195_730 |
0.2406859964 |
- |
- |
PREDICTED: protein LYK5-like [Jatropha curcas] |
| 18 |
Hb_000685_130 |
0.2452032331 |
- |
- |
PREDICTED: uncharacterized protein LOC105351820 [Fragaria vesca subsp. vesca] |
| 19 |
Hb_011716_050 |
0.2463799914 |
- |
- |
serine acetyltransferase [Hevea brasiliensis] |
| 20 |
Hb_000126_040 |
0.2500836138 |
- |
- |
kinase, putative [Ricinus communis] |