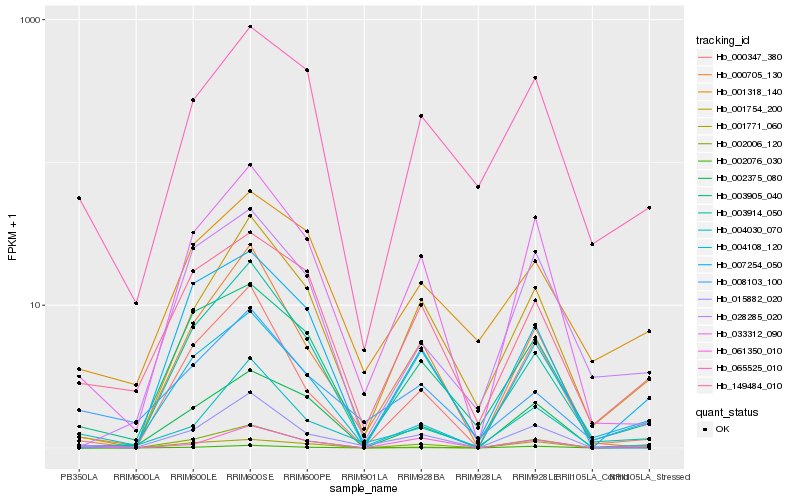
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002006_120 |
0.0 |
- |
- |
PREDICTED: glycine--tRNA ligase 1, mitochondrial-like [Jatropha curcas] |
| 2 |
Hb_003905_040 |
0.1956782898 |
- |
- |
PREDICTED: heptahelical transmembrane protein 2 [Jatropha curcas] |
| 3 |
Hb_007254_050 |
0.1983146683 |
- |
- |
PREDICTED: U-box domain-containing protein 28-like [Jatropha curcas] |
| 4 |
Hb_033312_090 |
0.1984124349 |
- |
- |
PREDICTED: HMG-Y-related protein A-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_149484_010 |
0.1986222494 |
transcription factor |
TF Family: NF-YC |
PREDICTED: dr1-associated corepressor-like [Eucalyptus grandis] |
| 6 |
Hb_061350_010 |
0.2000369665 |
- |
- |
PREDICTED: disease resistance protein RPS6-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_004030_070 |
0.2026370232 |
- |
- |
PREDICTED: uncharacterized protein LOC105632434 [Jatropha curcas] |
| 8 |
Hb_001754_200 |
0.2031277162 |
- |
- |
PREDICTED: U1 small nuclear ribonucleoprotein C [Jatropha curcas] |
| 9 |
Hb_000347_380 |
0.2033293951 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 10 |
Hb_065525_010 |
0.2037553256 |
- |
- |
hypothetical protein JCGZ_01481 [Jatropha curcas] |
| 11 |
Hb_015882_020 |
0.210940875 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 12 |
Hb_004108_120 |
0.2156397802 |
- |
- |
PREDICTED: EID1-like F-box protein 3 [Jatropha curcas] |
| 13 |
Hb_008103_100 |
0.2158272896 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPS2 [Jatropha curcas] |
| 14 |
Hb_001771_060 |
0.2200912486 |
- |
- |
hypothetical protein B456_012G144100 [Gossypium raimondii] |
| 15 |
Hb_002076_030 |
0.2204968861 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 16 |
Hb_028285_020 |
0.2288686822 |
transcription factor |
TF Family: C2C2-CO-like |
zinc finger protein, putative [Ricinus communis] |
| 17 |
Hb_002375_080 |
0.2294754518 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000705_130 |
0.2306113235 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001318_140 |
0.2306338953 |
- |
- |
PREDICTED: probable protein phosphatase 2C 49 [Jatropha curcas] |
| 20 |
Hb_003914_050 |
0.2309050349 |
- |
- |
PREDICTED: uncharacterized protein LOC104445137 [Eucalyptus grandis] |