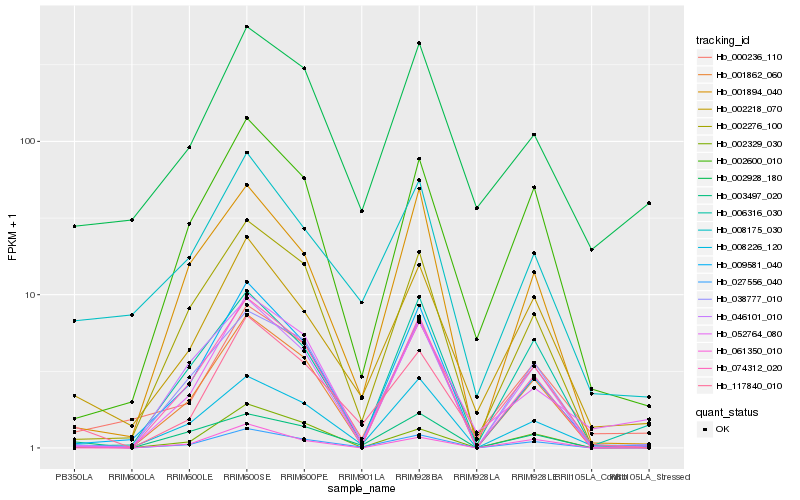
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027556_040 |
0.0 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Vitis vinifera] |
| 2 |
Hb_061350_010 |
0.1304060881 |
- |
- |
PREDICTED: disease resistance protein RPS6-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_002218_070 |
0.1435083871 |
- |
- |
PREDICTED: myosin-binding protein 1 [Jatropha curcas] |
| 4 |
Hb_006316_030 |
0.1583120442 |
- |
- |
aldehyde oxidase, putative [Ricinus communis] |
| 5 |
Hb_117840_010 |
0.162223396 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 6 |
Hb_038777_010 |
0.1662121613 |
transcription factor |
TF Family: G2-like |
PREDICTED: probable transcription factor KAN2 isoform X1 [Populus euphratica] |
| 7 |
Hb_003497_020 |
0.1686199063 |
transcription factor |
TF Family: MYB |
MYB transcriptional factor [Populus nigra] |
| 8 |
Hb_008226_120 |
0.1687392343 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 9 |
Hb_000236_110 |
0.1716645863 |
- |
- |
PREDICTED: chaperone protein dnaJ 20, chloroplastic [Jatropha curcas] |
| 10 |
Hb_009581_040 |
0.1742512172 |
- |
- |
hypothetical protein B456_011G052100 [Gossypium raimondii] |
| 11 |
Hb_002329_030 |
0.1761714228 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 2-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_001862_060 |
0.1782615203 |
- |
- |
PREDICTED: uncharacterized protein LOC105639802 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002928_180 |
0.1798015228 |
- |
- |
catalase CAT1 [Manihot esculenta] |
| 14 |
Hb_002600_010 |
0.1800805381 |
- |
- |
Allene oxide cyclase 4, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_002276_100 |
0.1806618981 |
- |
- |
PREDICTED: UDP-glycosyltransferase 89A2-like [Jatropha curcas] |
| 16 |
Hb_008175_030 |
0.1821098094 |
transcription factor |
TF Family: TAZ |
PREDICTED: BTB/POZ and TAZ domain-containing protein 3 [Jatropha curcas] |
| 17 |
Hb_052764_080 |
0.1827783235 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD5 isoform X1 [Jatropha curcas] |
| 18 |
Hb_074312_020 |
0.1857677147 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 19 |
Hb_001894_040 |
0.1862684623 |
- |
- |
PREDICTED: carbonic anhydrase 2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_046101_010 |
0.1867949217 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase-like protein CCR3 [Jatropha curcas] |