| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
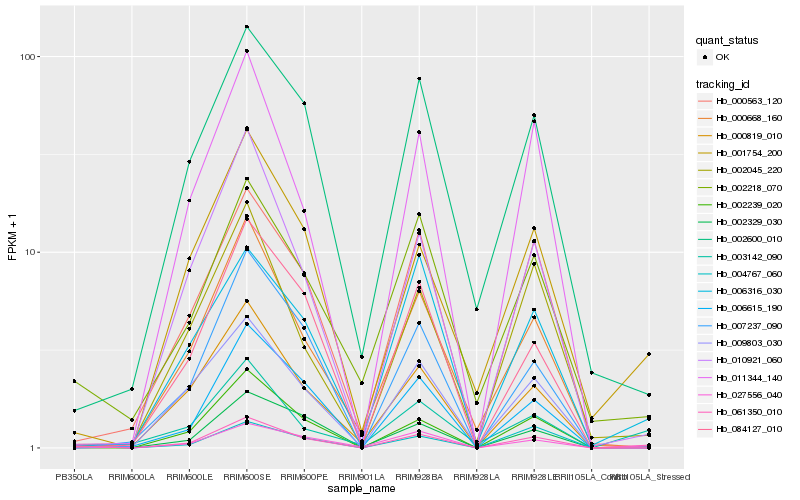
Hb_061350_010 |
0.0 |
- |
- |
PREDICTED: disease resistance protein RPS6-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_027556_040 |
0.1304060881 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Vitis vinifera] |
| 3 |
Hb_001754_200 |
0.1542083051 |
- |
- |
PREDICTED: U1 small nuclear ribonucleoprotein C [Jatropha curcas] |
| 4 |
Hb_004767_060 |
0.1639518409 |
- |
- |
PREDICTED: uncharacterized protein LOC105761928 [Gossypium raimondii] |
| 5 |
Hb_002329_030 |
0.1716676802 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 2-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_003142_090 |
0.1743839423 |
- |
- |
Kinase, putative isoform 2 [Theobroma cacao] |
| 7 |
Hb_000819_010 |
0.1750085485 |
- |
- |
Receptor-like protein kinase 1, putative [Theobroma cacao] |
| 8 |
Hb_010921_060 |
0.1762279159 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 9 |
Hb_002239_020 |
0.1762894611 |
- |
- |
MLO, putative [Theobroma cacao] |
| 10 |
Hb_006316_030 |
0.1795829612 |
- |
- |
aldehyde oxidase, putative [Ricinus communis] |
| 11 |
Hb_009803_030 |
0.1834017547 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Jatropha curcas] |
| 12 |
Hb_002218_070 |
0.184466767 |
- |
- |
PREDICTED: myosin-binding protein 1 [Jatropha curcas] |
| 13 |
Hb_002045_220 |
0.1856467199 |
- |
- |
PREDICTED: uncharacterized protein LOC105649919 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002600_010 |
0.1866804684 |
- |
- |
Allene oxide cyclase 4, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_000563_120 |
0.187711099 |
- |
- |
hypothetical protein JCGZ_18936 [Jatropha curcas] |
| 16 |
Hb_006615_190 |
0.1885528564 |
- |
- |
UDP-glycosyltransferase 1 [Linum usitatissimum] |
| 17 |
Hb_007237_090 |
0.1889201479 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000668_160 |
0.191022841 |
- |
- |
PREDICTED: sulfate transporter 1.3-like [Jatropha curcas] |
| 19 |
Hb_084127_010 |
0.1910242216 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 20 |
Hb_011344_140 |
0.1913313771 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like [Jatropha curcas] |