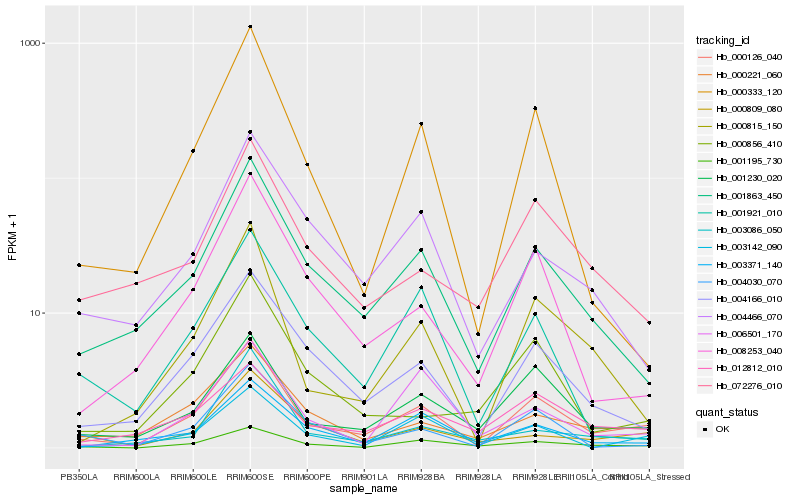
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000126_040 |
0.0 |
- |
- |
kinase, putative [Ricinus communis] |
| 2 |
Hb_001230_020 |
0.1475528721 |
- |
- |
hypothetical protein POPTR_0001s31610g [Populus trichocarpa] |
| 3 |
Hb_001863_450 |
0.1507095348 |
- |
- |
cryptochrome 1.2 [Populus tremula] |
| 4 |
Hb_004166_010 |
0.1580431501 |
- |
- |
PREDICTED: uncharacterized protein LOC105638508 [Jatropha curcas] |
| 5 |
Hb_000815_150 |
0.1635790685 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 6 |
Hb_001195_730 |
0.1652848227 |
- |
- |
PREDICTED: protein LYK5-like [Jatropha curcas] |
| 7 |
Hb_000221_060 |
0.175645115 |
- |
- |
- |
| 8 |
Hb_000333_120 |
0.1810172143 |
- |
- |
PREDICTED: thiol protease aleurain-like [Jatropha curcas] |
| 9 |
Hb_003142_090 |
0.1841318099 |
- |
- |
Kinase, putative isoform 2 [Theobroma cacao] |
| 10 |
Hb_008253_040 |
0.1853052668 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 11 |
Hb_012812_010 |
0.1862595818 |
- |
- |
putative leucine-rich repeat receptor-like serine/threonine-protein kinase [Morus notabilis] |
| 12 |
Hb_003371_140 |
0.1867316079 |
- |
- |
hypothetical protein POPTR_0001s23540g, partial [Populus trichocarpa] |
| 13 |
Hb_004030_070 |
0.189560444 |
- |
- |
PREDICTED: uncharacterized protein LOC105632434 [Jatropha curcas] |
| 14 |
Hb_072276_010 |
0.195170803 |
- |
- |
PREDICTED: elongation of fatty acids protein 3-like [Jatropha curcas] |
| 15 |
Hb_004466_070 |
0.1962170562 |
- |
- |
hypothetical protein RCOM_1447500 [Ricinus communis] |
| 16 |
Hb_000809_080 |
0.1996250239 |
- |
- |
50S ribosomal protein L25, putative [Ricinus communis] |
| 17 |
Hb_001921_010 |
0.2048690824 |
- |
- |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 18 |
Hb_003086_050 |
0.2072773379 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 19 |
Hb_006501_170 |
0.2075009531 |
- |
- |
hypothetical protein JCGZ_07371 [Jatropha curcas] |
| 20 |
Hb_000856_410 |
0.2096008032 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |