| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
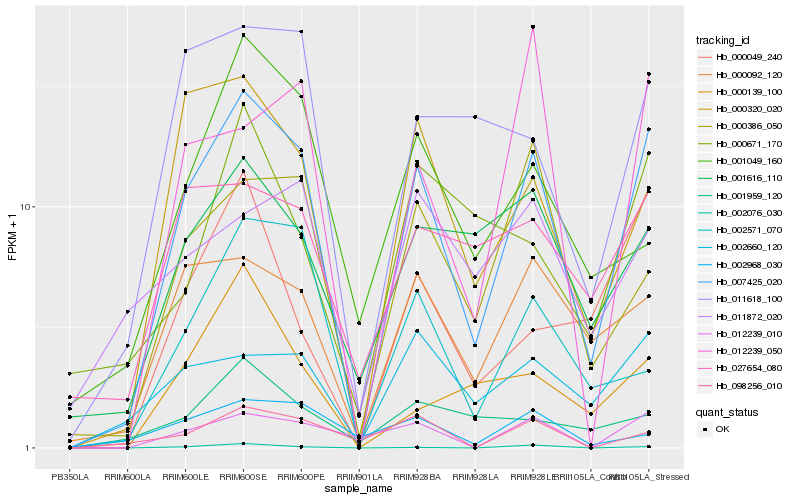
Hb_007425_020 |
0.0 |
transcription factor |
TF Family: ERF |
PREDICTED: pathogenesis-related genes transcriptional activator PTI6-like [Jatropha curcas] |
| 2 |
Hb_000092_120 |
0.1991971994 |
- |
- |
PREDICTED: non-structural maintenance of chromosomes element 4 homolog A-like [Populus euphratica] |
| 3 |
Hb_000320_020 |
0.2017964723 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 31 [Jatropha curcas] |
| 4 |
Hb_012239_010 |
0.2140204598 |
- |
- |
- |
| 5 |
Hb_000386_050 |
0.2174556891 |
- |
- |
PREDICTED: 2-isopropylmalate synthase 2, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_002660_120 |
0.2194988504 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase [Populus euphratica] |
| 7 |
Hb_011618_100 |
0.2263593463 |
transcription factor |
TF Family: AUX/IAA |
Auxin-responsive protein IAA1, putative [Ricinus communis] |
| 8 |
Hb_001616_110 |
0.2279672443 |
- |
- |
PREDICTED: RPM1-interacting protein 4 [Jatropha curcas] |
| 9 |
Hb_002571_070 |
0.2339583188 |
- |
- |
PREDICTED: cation/calcium exchanger 2 [Jatropha curcas] |
| 10 |
Hb_002076_030 |
0.2347921958 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 11 |
Hb_012239_050 |
0.2381984915 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_027654_080 |
0.2401213204 |
- |
- |
PREDICTED: carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000049_240 |
0.2416209733 |
- |
- |
PREDICTED: uncharacterized protein LOC105644456 [Jatropha curcas] |
| 14 |
Hb_098256_010 |
0.2425985815 |
- |
- |
PREDICTED: uncharacterized protein LOC105801076 [Gossypium raimondii] |
| 15 |
Hb_001049_160 |
0.2489212099 |
- |
- |
aldose 1-epimerase, putative [Ricinus communis] |
| 16 |
Hb_000139_100 |
0.2514991334 |
- |
- |
PREDICTED: sugar transport protein 13-like [Jatropha curcas] |
| 17 |
Hb_002968_030 |
0.2525401669 |
- |
- |
Pectinesterase-1 precursor, putative [Ricinus communis] |
| 18 |
Hb_001959_120 |
0.2539007718 |
- |
- |
PREDICTED: uncharacterized protein LOC105638487 [Jatropha curcas] |
| 19 |
Hb_011872_020 |
0.2567677165 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH128 [Jatropha curcas] |
| 20 |
Hb_000671_170 |
0.2580497831 |
- |
- |
kinase, putative [Ricinus communis] |