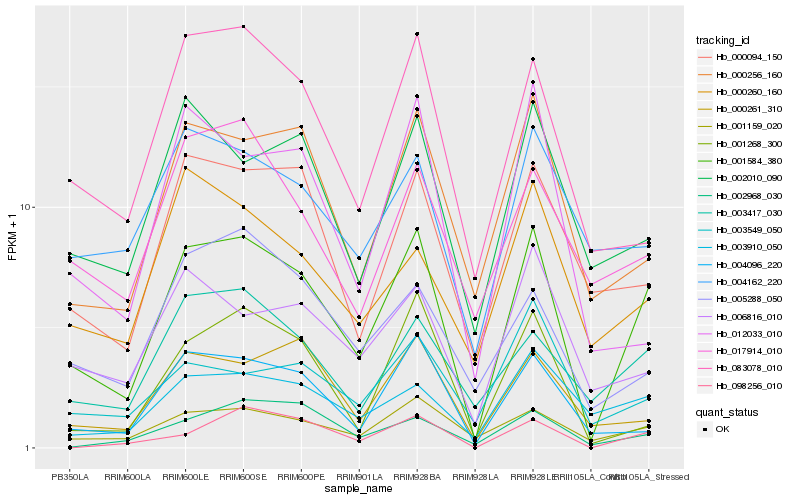
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001584_380 |
0.0 |
- |
- |
PREDICTED: tropomyosin-like [Jatropha curcas] |
| 2 |
Hb_001159_020 |
0.1637321887 |
- |
- |
- |
| 3 |
Hb_000256_160 |
0.1756019443 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein knotted-1-like 3-like isoform X1 [Glycine max] |
| 4 |
Hb_004096_220 |
0.1756410048 |
- |
- |
PREDICTED: uncharacterized protein LOC105638706 isoform X1 [Jatropha curcas] |
| 5 |
Hb_003417_030 |
0.1850124964 |
- |
- |
PREDICTED: sister chromatid cohesion 1 protein 2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000094_150 |
0.1852174261 |
- |
- |
- |
| 7 |
Hb_000261_310 |
0.1854339616 |
- |
- |
PREDICTED: UV radiation resistance-associated gene protein isoform X3 [Jatropha curcas] |
| 8 |
Hb_083078_010 |
0.1859680098 |
- |
- |
Splicing factor, arginine/serine-rich, putative [Ricinus communis] |
| 9 |
Hb_003910_050 |
0.1864158658 |
- |
- |
PREDICTED: uncharacterized protein LOC105646926 [Jatropha curcas] |
| 10 |
Hb_002010_090 |
0.1873425339 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |
| 11 |
Hb_006816_010 |
0.1899469649 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001268_300 |
0.190820281 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_012033_010 |
0.1978675546 |
- |
- |
PREDICTED: histone H1.2-like [Jatropha curcas] |
| 14 |
Hb_003549_050 |
0.1980418418 |
- |
- |
PREDICTED: zinc phosphodiesterase ELAC protein 2-like [Jatropha curcas] |
| 15 |
Hb_098256_010 |
0.2001947237 |
- |
- |
PREDICTED: uncharacterized protein LOC105801076 [Gossypium raimondii] |
| 16 |
Hb_000260_160 |
0.2008855986 |
transcription factor |
TF Family: GNAT |
ATP binding protein, putative [Ricinus communis] |
| 17 |
Hb_002968_030 |
0.2021802606 |
- |
- |
Pectinesterase-1 precursor, putative [Ricinus communis] |
| 18 |
Hb_004162_220 |
0.2030275685 |
- |
- |
PREDICTED: pheophorbide a oxygenase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_005288_050 |
0.2039448165 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase SUVR4 [Jatropha curcas] |
| 20 |
Hb_017914_010 |
0.2045364855 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SUPPRESSOR OF FRI 4 [Jatropha curcas] |