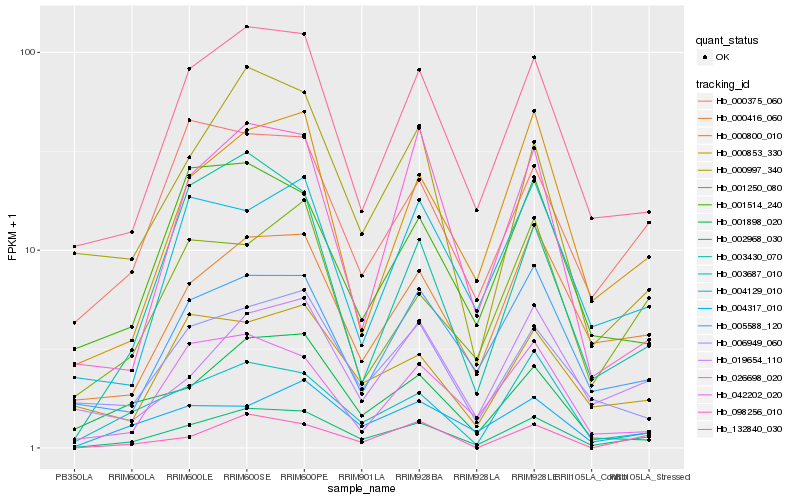
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002968_030 |
0.0 |
- |
- |
Pectinesterase-1 precursor, putative [Ricinus communis] |
| 2 |
Hb_132840_030 |
0.1161047752 |
transcription factor |
TF Family: MYB-related |
lhy1 [Populus tremula] |
| 3 |
Hb_000416_060 |
0.1317278218 |
- |
- |
PREDICTED: uncharacterized protein LOC105649718 [Jatropha curcas] |
| 4 |
Hb_000800_010 |
0.1414210789 |
- |
- |
PREDICTED: glycosyltransferase family protein 64 protein C5 [Jatropha curcas] |
| 5 |
Hb_003687_010 |
0.1456494108 |
- |
- |
Cysteine-rich RLK 29, putative [Theobroma cacao] |
| 6 |
Hb_005588_120 |
0.146487957 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000375_060 |
0.1538158905 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1-like [Jatropha curcas] |
| 8 |
Hb_004317_010 |
0.1546022242 |
- |
- |
hypothetical protein CICLE_v10011038mg [Citrus clementina] |
| 9 |
Hb_000997_340 |
0.1565810807 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 10 |
Hb_019654_110 |
0.1569275622 |
- |
- |
PREDICTED: F-box protein SKIP23-like [Jatropha curcas] |
| 11 |
Hb_006949_060 |
0.1587401608 |
- |
- |
PREDICTED: aminopeptidase M1-like [Jatropha curcas] |
| 12 |
Hb_001250_080 |
0.1613041902 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g23390 [Jatropha curcas] |
| 13 |
Hb_098256_010 |
0.1635114281 |
- |
- |
PREDICTED: uncharacterized protein LOC105801076 [Gossypium raimondii] |
| 14 |
Hb_026698_020 |
0.1647753571 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 15 |
Hb_001514_240 |
0.1648438417 |
- |
- |
PREDICTED: aspartic proteinase CDR1 [Jatropha curcas] |
| 16 |
Hb_042202_020 |
0.1651191769 |
- |
- |
Eukaryotic release factor 1-3 isoform 1 [Theobroma cacao] |
| 17 |
Hb_004129_010 |
0.1665391866 |
- |
- |
aspartate aminotransferase, putative [Ricinus communis] |
| 18 |
Hb_001898_020 |
0.1668056774 |
- |
- |
PREDICTED: uncharacterized protein LOC105629659 [Jatropha curcas] |
| 19 |
Hb_000853_330 |
0.1672562916 |
- |
- |
PREDICTED: importin subunit alpha-like [Jatropha curcas] |
| 20 |
Hb_003430_070 |
0.1676033379 |
- |
- |
PREDICTED: protein phosphatase 2C 37 [Jatropha curcas] |