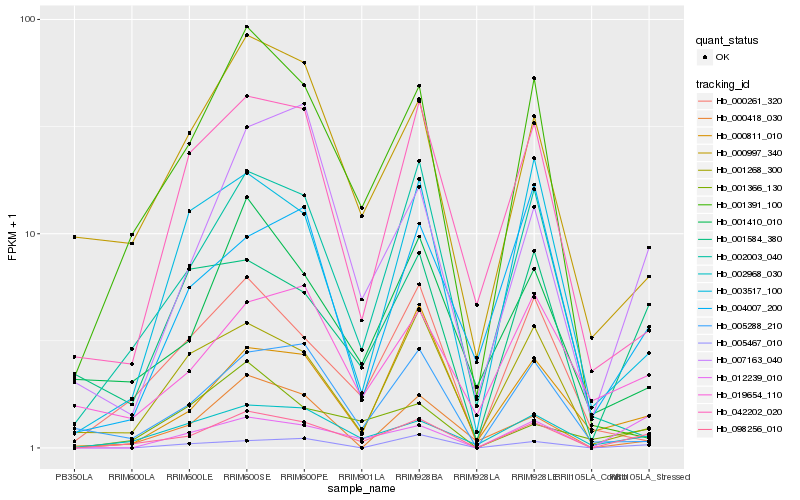
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_098256_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105801076 [Gossypium raimondii] |
| 2 |
Hb_002968_030 |
0.1635114281 |
- |
- |
Pectinesterase-1 precursor, putative [Ricinus communis] |
| 3 |
Hb_012239_010 |
0.1983302886 |
- |
- |
- |
| 4 |
Hb_001584_380 |
0.2001947237 |
- |
- |
PREDICTED: tropomyosin-like [Jatropha curcas] |
| 5 |
Hb_007163_040 |
0.2155600431 |
- |
- |
PREDICTED: uncharacterized protein At5g65660 [Vitis vinifera] |
| 6 |
Hb_003517_100 |
0.2158469909 |
transcription factor |
TF Family: GRAS |
- |
| 7 |
Hb_001268_300 |
0.2188800889 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000997_340 |
0.2210295293 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 9 |
Hb_001391_100 |
0.2226574668 |
- |
- |
AMP dependent ligase, putative [Ricinus communis] |
| 10 |
Hb_001410_010 |
0.2258595433 |
transcription factor |
TF Family: RWP-RK |
PREDICTED: protein NLP7-like [Jatropha curcas] |
| 11 |
Hb_042202_020 |
0.2259808918 |
- |
- |
Eukaryotic release factor 1-3 isoform 1 [Theobroma cacao] |
| 12 |
Hb_000811_010 |
0.2262189375 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF118 [Jatropha curcas] |
| 13 |
Hb_005467_010 |
0.2264926627 |
transcription factor |
TF Family: ERF |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_019654_110 |
0.2276564288 |
- |
- |
PREDICTED: F-box protein SKIP23-like [Jatropha curcas] |
| 15 |
Hb_002003_040 |
0.2280480994 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000261_320 |
0.2288716742 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_000418_030 |
0.2295854798 |
- |
- |
PREDICTED: uncharacterized protein LOC105636893 [Jatropha curcas] |
| 18 |
Hb_005288_210 |
0.2303974303 |
- |
- |
hypothetical protein VITISV_017217 [Vitis vinifera] |
| 19 |
Hb_001366_130 |
0.2306892288 |
- |
- |
unnamed protein product [Coffea canephora] |
| 20 |
Hb_004007_200 |
0.2313611926 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein NPY5 [Jatropha curcas] |