| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
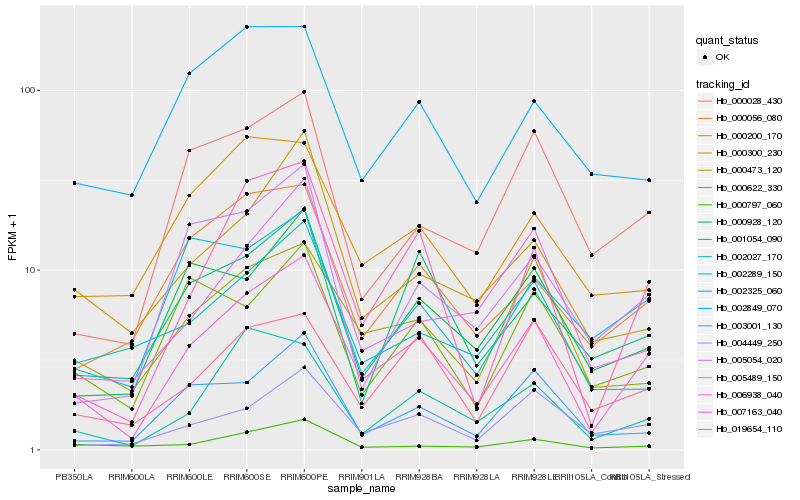
Hb_006938_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At1g66480-like [Populus euphratica] |
| 2 |
Hb_007163_040 |
0.1380309123 |
- |
- |
PREDICTED: uncharacterized protein At5g65660 [Vitis vinifera] |
| 3 |
Hb_004449_250 |
0.1615610573 |
- |
- |
PREDICTED: protein EI24 homolog [Jatropha curcas] |
| 4 |
Hb_000056_080 |
0.1656244156 |
- |
- |
PREDICTED: protein kinase 2B, chloroplastic [Jatropha curcas] |
| 5 |
Hb_002027_170 |
0.1674627937 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RKF3 [Jatropha curcas] |
| 6 |
Hb_000473_120 |
0.169035784 |
- |
- |
hypothetical protein JCGZ_16177 [Jatropha curcas] |
| 7 |
Hb_000928_120 |
0.1699778449 |
- |
- |
PREDICTED: benzyl alcohol O-benzoyltransferase-like [Jatropha curcas] |
| 8 |
Hb_000028_430 |
0.1700157411 |
- |
- |
PREDICTED: uncharacterized protein LOC105641369 isoform X1 [Jatropha curcas] |
| 9 |
Hb_005054_020 |
0.1715318054 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g11320 [Jatropha curcas] |
| 10 |
Hb_003001_130 |
0.1718783979 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_019654_110 |
0.1741658924 |
- |
- |
PREDICTED: F-box protein SKIP23-like [Jatropha curcas] |
| 12 |
Hb_000300_230 |
0.1772686813 |
- |
- |
PREDICTED: cation/calcium exchanger 2 [Jatropha curcas] |
| 13 |
Hb_001054_090 |
0.1777190795 |
- |
- |
Potassium channel SKOR, putative [Ricinus communis] |
| 14 |
Hb_000200_170 |
0.1790073899 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA2A-like [Populus euphratica] |
| 15 |
Hb_002849_070 |
0.1810728304 |
- |
- |
PREDICTED: protein BPS1, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000622_330 |
0.183860735 |
- |
- |
PREDICTED: uncharacterized protein LOC105642906 [Jatropha curcas] |
| 17 |
Hb_005489_150 |
0.1842073256 |
- |
- |
hypothetical protein JCGZ_20759 [Jatropha curcas] |
| 18 |
Hb_000797_060 |
0.1847653453 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 19 |
Hb_002325_060 |
0.1849130086 |
- |
- |
PREDICTED: ADP-ribosylation factor-like protein 8B-like [Glycine max] |
| 20 |
Hb_002289_150 |
0.1862497787 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 3 [UDP-forming]-like [Jatropha curcas] |