| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
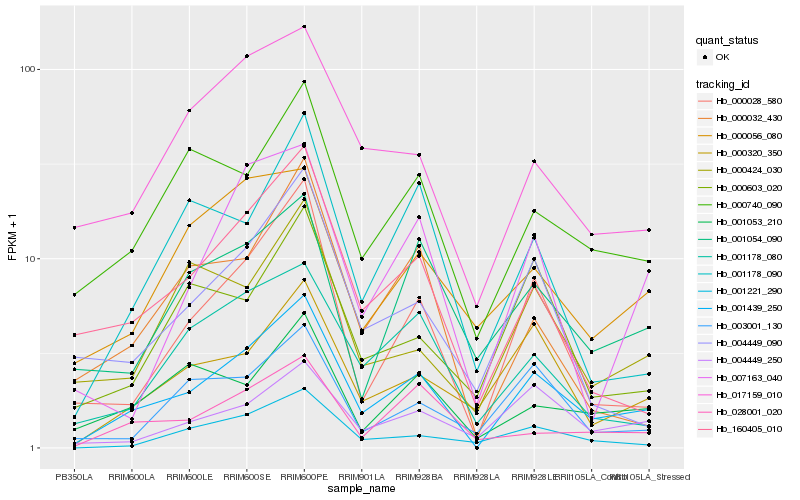
Hb_001439_250 |
0.0 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 100 [Jatropha curcas] |
| 2 |
Hb_000028_580 |
0.1605630531 |
- |
- |
PREDICTED: glutamate decarboxylase 5-like [Jatropha curcas] |
| 3 |
Hb_000320_350 |
0.1790432243 |
- |
- |
hypothetical protein RCOM_0904330 [Ricinus communis] |
| 4 |
Hb_000740_090 |
0.1791539792 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_160405_010 |
0.1803556648 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 6 |
Hb_001178_080 |
0.1876444417 |
- |
- |
PREDICTED: protein kinase PVPK-1 [Jatropha curcas] |
| 7 |
Hb_004449_250 |
0.1876693774 |
- |
- |
PREDICTED: protein EI24 homolog [Jatropha curcas] |
| 8 |
Hb_017159_010 |
0.1877763959 |
- |
- |
- |
| 9 |
Hb_001221_290 |
0.188618634 |
- |
- |
hypothetical protein RCOM_1506010 [Ricinus communis] |
| 10 |
Hb_004449_090 |
0.1957048573 |
- |
- |
PREDICTED: CO(2)-response secreted protease-like [Jatropha curcas] |
| 11 |
Hb_007163_040 |
0.1960240406 |
- |
- |
PREDICTED: uncharacterized protein At5g65660 [Vitis vinifera] |
| 12 |
Hb_028001_020 |
0.1963030029 |
- |
- |
PREDICTED: lysM domain receptor-like kinase 4 [Jatropha curcas] |
| 13 |
Hb_001178_090 |
0.1976950626 |
- |
- |
Monodehydroascorbate reductase family protein [Populus trichocarpa] |
| 14 |
Hb_003001_130 |
0.1985259274 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001053_210 |
0.1994502092 |
- |
- |
- |
| 16 |
Hb_001054_090 |
0.1999901078 |
- |
- |
Potassium channel SKOR, putative [Ricinus communis] |
| 17 |
Hb_000032_430 |
0.2007070884 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000424_030 |
0.2025666542 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000056_080 |
0.2060200637 |
- |
- |
PREDICTED: protein kinase 2B, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000603_020 |
0.2060434585 |
- |
- |
PREDICTED: uncharacterized protein LOC105638063 isoform X1 [Jatropha curcas] |