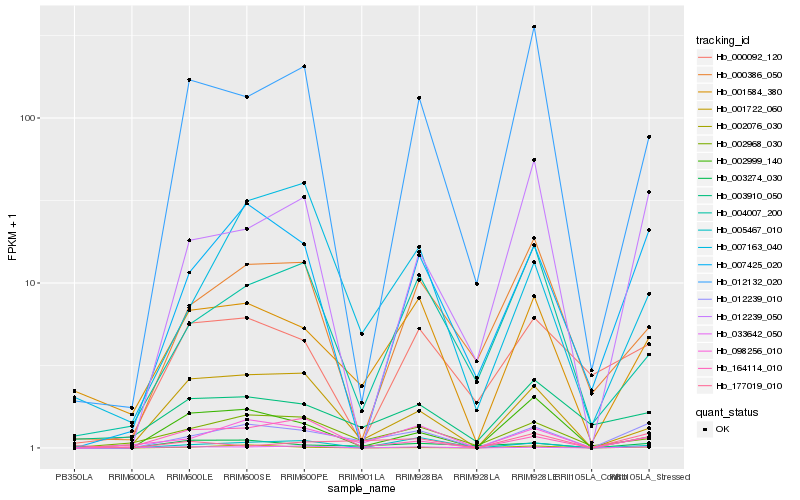
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012239_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_098256_010 |
0.1983302886 |
- |
- |
PREDICTED: uncharacterized protein LOC105801076 [Gossypium raimondii] |
| 3 |
Hb_007425_020 |
0.2140204598 |
transcription factor |
TF Family: ERF |
PREDICTED: pathogenesis-related genes transcriptional activator PTI6-like [Jatropha curcas] |
| 4 |
Hb_001584_380 |
0.2291335501 |
- |
- |
PREDICTED: tropomyosin-like [Jatropha curcas] |
| 5 |
Hb_012239_050 |
0.2504586813 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002968_030 |
0.26441063 |
- |
- |
Pectinesterase-1 precursor, putative [Ricinus communis] |
| 7 |
Hb_003910_050 |
0.2855949009 |
- |
- |
PREDICTED: uncharacterized protein LOC105646926 [Jatropha curcas] |
| 8 |
Hb_164114_010 |
0.2867891343 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 9 |
Hb_002076_030 |
0.2870914815 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 10 |
Hb_007163_040 |
0.2886297774 |
- |
- |
PREDICTED: uncharacterized protein At5g65660 [Vitis vinifera] |
| 11 |
Hb_005467_010 |
0.2904283863 |
transcription factor |
TF Family: ERF |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_177019_010 |
0.2942744997 |
- |
- |
Uncharacterized protein TCM_004012 [Theobroma cacao] |
| 13 |
Hb_001722_060 |
0.2946695739 |
- |
- |
- |
| 14 |
Hb_000386_050 |
0.2956399492 |
- |
- |
PREDICTED: 2-isopropylmalate synthase 2, chloroplastic-like [Jatropha curcas] |
| 15 |
Hb_003274_030 |
0.296734807 |
- |
- |
putative wall-associated kinase family protein [Populus trichocarpa] |
| 16 |
Hb_033642_050 |
0.2975619337 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter B family member 4-like [Jatropha curcas] |
| 17 |
Hb_012132_020 |
0.2980463118 |
- |
- |
glutamine synthetase [Hevea brasiliensis] |
| 18 |
Hb_002999_140 |
0.298713306 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 19 |
Hb_000092_120 |
0.2987617919 |
- |
- |
PREDICTED: non-structural maintenance of chromosomes element 4 homolog A-like [Populus euphratica] |
| 20 |
Hb_004007_200 |
0.2994668978 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein NPY5 [Jatropha curcas] |