| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
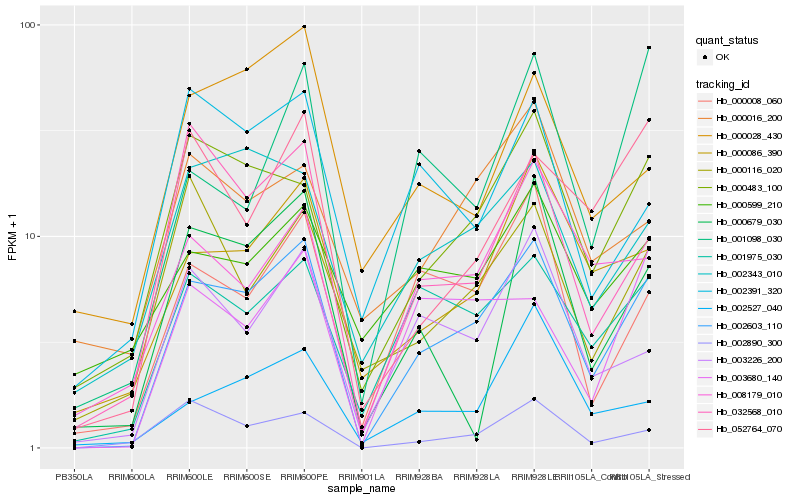
Hb_002603_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105646325 [Jatropha curcas] |
| 2 |
Hb_001975_030 |
0.1660664267 |
- |
- |
4-diphosphocytidyl-2-c-methyl-d-erythritol kinase, partial [Plectranthus barbatus] |
| 3 |
Hb_000483_100 |
0.1739861407 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 9-like [Jatropha curcas] |
| 4 |
Hb_000008_060 |
0.1759855564 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 5 |
Hb_000086_390 |
0.1801497416 |
- |
- |
PREDICTED: uncharacterized protein LOC105644503 [Jatropha curcas] |
| 6 |
Hb_003226_200 |
0.1811727098 |
- |
- |
magnesium/proton exchanger, putative [Ricinus communis] |
| 7 |
Hb_032568_010 |
0.1857802049 |
- |
- |
PREDICTED: protein phosphatase 2C 56-like [Jatropha curcas] |
| 8 |
Hb_003680_140 |
0.1897792874 |
- |
- |
PREDICTED: uncharacterized protein LOC105121770 [Populus euphratica] |
| 9 |
Hb_002343_010 |
0.1915353537 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 10 |
Hb_008179_010 |
0.1919501237 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 16 [Jatropha curcas] |
| 11 |
Hb_002890_300 |
0.1934640677 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 12 |
Hb_052764_070 |
0.1980458949 |
- |
- |
PREDICTED: uncharacterized protein LOC105649473 [Jatropha curcas] |
| 13 |
Hb_000116_020 |
0.2061788882 |
- |
- |
PREDICTED: serine acetyltransferase 2-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_002391_320 |
0.2078587735 |
- |
- |
PREDICTED: phospholipase D delta-like [Jatropha curcas] |
| 15 |
Hb_001098_030 |
0.2087192679 |
- |
- |
PREDICTED: probable NAD(P)H dehydrogenase (quinone) FQR1-like 1 [Populus euphratica] |
| 16 |
Hb_002527_040 |
0.2103653574 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat-containing protein 2, lrrc2, putative [Ricinus communis] |
| 17 |
Hb_000599_210 |
0.2118783958 |
- |
- |
serine/threonine-protein kinase, putative [Ricinus communis] |
| 18 |
Hb_000028_430 |
0.213276129 |
- |
- |
PREDICTED: uncharacterized protein LOC105641369 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000016_200 |
0.2133840261 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000679_030 |
0.2135590876 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HY5 [Jatropha curcas] |