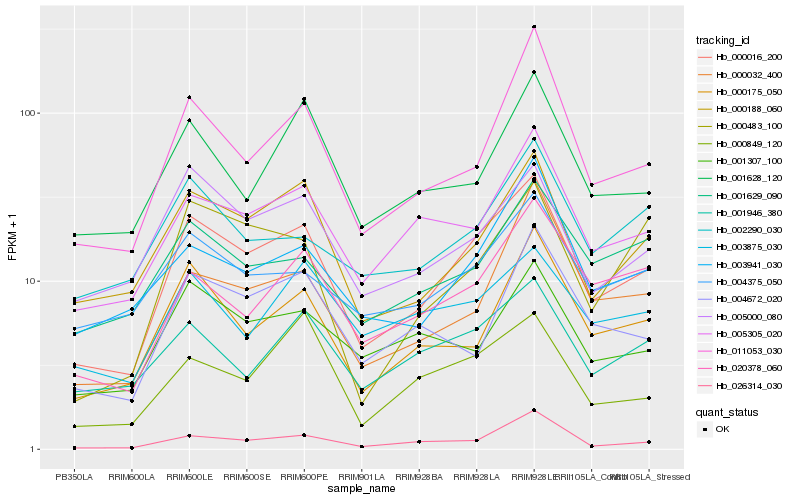
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000016_200 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000188_060 |
0.1163823277 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 4-like [Jatropha curcas] |
| 3 |
Hb_005305_020 |
0.1279214596 |
- |
- |
PREDICTED: dicarboxylate transporter 1, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000175_050 |
0.134355396 |
- |
- |
- |
| 5 |
Hb_011053_030 |
0.1344687783 |
- |
- |
- |
| 6 |
Hb_005000_080 |
0.136208999 |
- |
- |
PREDICTED: thioredoxin-like 2, chloroplastic [Populus euphratica] |
| 7 |
Hb_020378_060 |
0.1404303871 |
- |
- |
PREDICTED: methyltransferase-like protein 13 [Jatropha curcas] |
| 8 |
Hb_026314_030 |
0.141631223 |
- |
- |
PREDICTED: transmembrane protein 41B [Jatropha curcas] |
| 9 |
Hb_003941_030 |
0.145217971 |
- |
- |
Protease ecfE, putative [Ricinus communis] |
| 10 |
Hb_004375_050 |
0.146519035 |
- |
- |
PREDICTED: putative GTP diphosphokinase RSH1, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_001946_380 |
0.1472245542 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Jatropha curcas] |
| 12 |
Hb_000032_400 |
0.1506169738 |
- |
- |
PREDICTED: UDP-glucose 4-epimerase GEPI48 [Jatropha curcas] |
| 13 |
Hb_001629_090 |
0.1519118195 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001307_100 |
0.1527858994 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 15 |
Hb_000849_120 |
0.1530062777 |
- |
- |
PREDICTED: linoleate 9S-lipoxygenase 6-like [Jatropha curcas] |
| 16 |
Hb_004672_020 |
0.1535607281 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 17 |
Hb_001628_120 |
0.1538772287 |
- |
- |
putative ascorbate peroxidase, partial [Taraxacum brevicorniculatum] |
| 18 |
Hb_000483_100 |
0.1561552591 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 9-like [Jatropha curcas] |
| 19 |
Hb_002290_030 |
0.1565267229 |
- |
- |
PREDICTED: uncharacterized protein LOC105635877 isoform X1 [Jatropha curcas] |
| 20 |
Hb_003875_030 |
0.158046943 |
- |
- |
PREDICTED: probable plastidic glucose transporter 1 [Jatropha curcas] |