| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
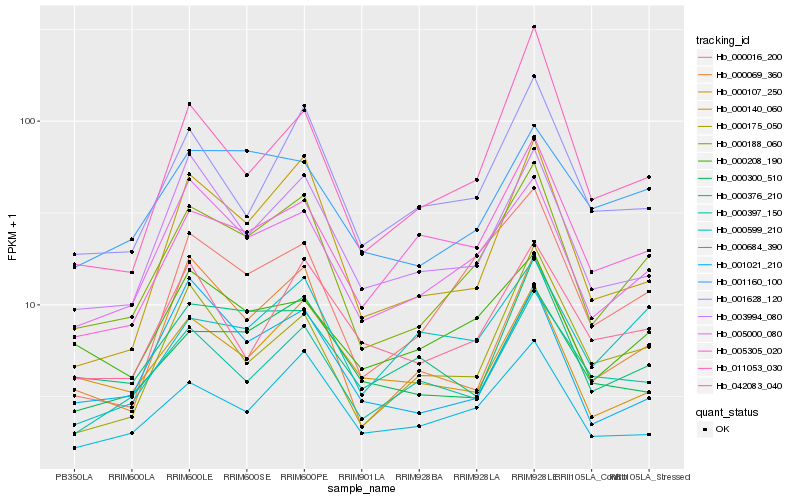
Hb_000188_060 |
0.0 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 4-like [Jatropha curcas] |
| 2 |
Hb_005000_080 |
0.0972643141 |
- |
- |
PREDICTED: thioredoxin-like 2, chloroplastic [Populus euphratica] |
| 3 |
Hb_000016_200 |
0.1163823277 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000684_390 |
0.1210425627 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 4 [Jatropha curcas] |
| 5 |
Hb_003994_080 |
0.1217393685 |
- |
- |
PREDICTED: uncharacterized protein LOC105634384 [Jatropha curcas] |
| 6 |
Hb_000140_060 |
0.1245200756 |
- |
- |
50S ribosomal protein L5, putative [Ricinus communis] |
| 7 |
Hb_000069_360 |
0.1251620171 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001628_120 |
0.1251915467 |
- |
- |
putative ascorbate peroxidase, partial [Taraxacum brevicorniculatum] |
| 9 |
Hb_000208_190 |
0.1316700899 |
- |
- |
PREDICTED: uncharacterized protein LOC105648580 [Jatropha curcas] |
| 10 |
Hb_001160_100 |
0.1368347163 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 6, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000376_210 |
0.1375431002 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like [Jatropha curcas] |
| 12 |
Hb_005305_020 |
0.1378034805 |
- |
- |
PREDICTED: dicarboxylate transporter 1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000397_150 |
0.1385740499 |
- |
- |
PREDICTED: probable flavin-containing monooxygenase 1 [Jatropha curcas] |
| 14 |
Hb_000300_510 |
0.1390959777 |
- |
- |
PREDICTED: uncharacterized protein LOC105632605 [Jatropha curcas] |
| 15 |
Hb_000175_050 |
0.1398193119 |
- |
- |
- |
| 16 |
Hb_000107_250 |
0.1439458966 |
- |
- |
voltage-gated clc-type chloride channel, putative [Ricinus communis] |
| 17 |
Hb_001021_210 |
0.1443735755 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_042083_040 |
0.1443943103 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_011053_030 |
0.1448278414 |
- |
- |
- |
| 20 |
Hb_000599_210 |
0.1451522036 |
- |
- |
serine/threonine-protein kinase, putative [Ricinus communis] |