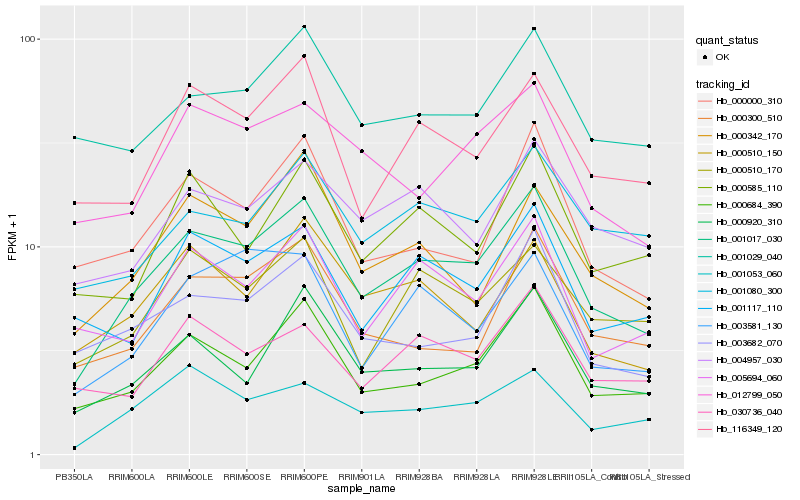
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001017_030 |
0.0 |
- |
- |
copine, putative [Ricinus communis] |
| 2 |
Hb_000684_390 |
0.0999150112 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 4 [Jatropha curcas] |
| 3 |
Hb_000920_310 |
0.1114299277 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 4 |
Hb_116349_120 |
0.114569345 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit alpha-1, mitochondrial isoform X1 [Jatropha curcas] |
| 5 |
Hb_001053_060 |
0.1174122138 |
- |
- |
DHHC-type zinc finger family protein [Theobroma cacao] |
| 6 |
Hb_003682_070 |
0.1188489837 |
- |
- |
PREDICTED: uncharacterized protein LOC105633817 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000342_170 |
0.1203450786 |
- |
- |
PREDICTED: uncharacterized protein LOC100807540 isoform X2 [Glycine max] |
| 8 |
Hb_001117_110 |
0.1209506458 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 9 |
Hb_005694_060 |
0.1209626126 |
- |
- |
PREDICTED: bifunctional fucokinase/fucose pyrophosphorylase [Jatropha curcas] |
| 10 |
Hb_000000_310 |
0.1209816964 |
- |
- |
metalloendopeptidase, putative [Ricinus communis] |
| 11 |
Hb_000510_170 |
0.1213037027 |
- |
- |
Glycosyltransferase QUASIMODO1, putative [Ricinus communis] |
| 12 |
Hb_004957_030 |
0.1217135472 |
- |
- |
hydroxyproline-rich glycoprotein [Populus trichocarpa] |
| 13 |
Hb_012799_050 |
0.1219959863 |
- |
- |
PREDICTED: remorin [Jatropha curcas] |
| 14 |
Hb_001080_300 |
0.1234041138 |
- |
- |
PREDICTED: F-box protein SKIP17-like [Jatropha curcas] |
| 15 |
Hb_000510_150 |
0.1240161958 |
- |
- |
PREDICTED: magnesium transporter MRS2-I-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_030736_040 |
0.1256959425 |
- |
- |
PREDICTED: uncharacterized protein LOC105638926 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000300_510 |
0.1260004959 |
- |
- |
PREDICTED: uncharacterized protein LOC105632605 [Jatropha curcas] |
| 18 |
Hb_000585_110 |
0.126211919 |
- |
- |
PREDICTED: probable protein phosphatase 2C 66 [Jatropha curcas] |
| 19 |
Hb_003581_130 |
0.1262881119 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_001029_040 |
0.1265282819 |
- |
- |
PREDICTED: uncharacterized protein LOC105641058 [Jatropha curcas] |