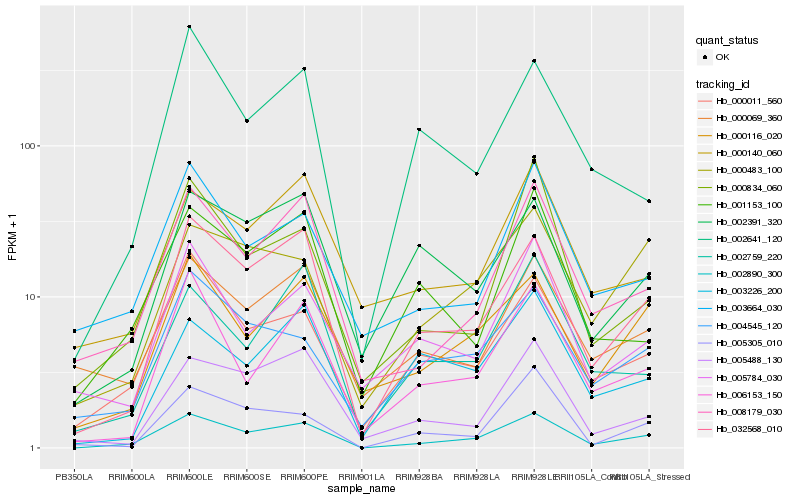
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002890_300 |
0.0 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 2 |
Hb_032568_010 |
0.1155811382 |
- |
- |
PREDICTED: protein phosphatase 2C 56-like [Jatropha curcas] |
| 3 |
Hb_008179_030 |
0.1495255374 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 8 [Jatropha curcas] |
| 4 |
Hb_000011_560 |
0.1504978355 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000116_020 |
0.1530291703 |
- |
- |
PREDICTED: serine acetyltransferase 2-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_006153_150 |
0.1654162621 |
- |
- |
PREDICTED: uncharacterized protein LOC105645999 [Jatropha curcas] |
| 7 |
Hb_000483_100 |
0.1690863216 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 9-like [Jatropha curcas] |
| 8 |
Hb_004545_120 |
0.170364239 |
- |
- |
PREDICTED: uncharacterized protein LOC105647604 [Jatropha curcas] |
| 9 |
Hb_003664_030 |
0.1756400608 |
- |
- |
PREDICTED: mitochondrial carnitine/acylcarnitine carrier-like protein [Jatropha curcas] |
| 10 |
Hb_000834_060 |
0.1785261781 |
- |
- |
PREDICTED: probable chlorophyll(ide) b reductase NYC1, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_002759_220 |
0.1800088002 |
- |
- |
altered response to gravity (arg1), plant, putative [Ricinus communis] |
| 12 |
Hb_000069_360 |
0.1800605381 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_005488_130 |
0.1828106543 |
- |
- |
transporter, putative [Ricinus communis] |
| 14 |
Hb_005305_010 |
0.1829666272 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB46 [Jatropha curcas] |
| 15 |
Hb_001153_100 |
0.1849540356 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein HD1 [Jatropha curcas] |
| 16 |
Hb_005784_030 |
0.1850108252 |
- |
- |
PREDICTED: uncharacterized protein LOC105645162 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000140_060 |
0.1858409174 |
- |
- |
50S ribosomal protein L5, putative [Ricinus communis] |
| 18 |
Hb_002391_320 |
0.1863791782 |
- |
- |
PREDICTED: phospholipase D delta-like [Jatropha curcas] |
| 19 |
Hb_003226_200 |
0.1870857203 |
- |
- |
magnesium/proton exchanger, putative [Ricinus communis] |
| 20 |
Hb_002641_120 |
0.1874566914 |
- |
- |
PREDICTED: calmodulin [Jatropha curcas] |