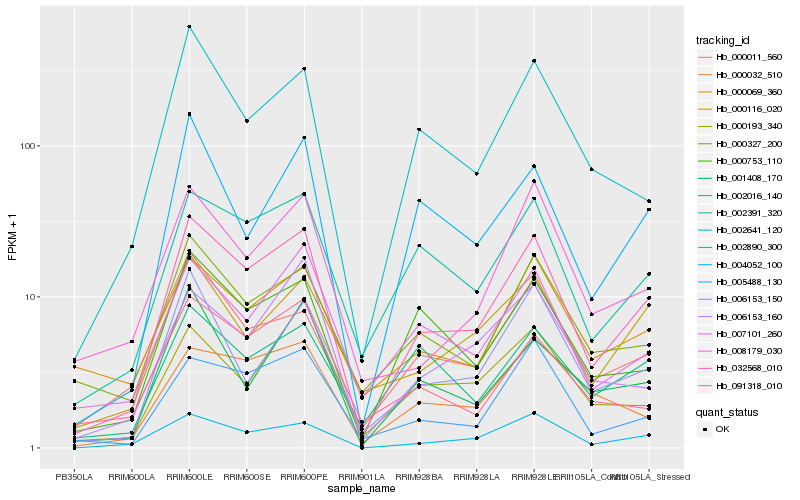
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_032568_010 |
0.0 |
- |
- |
PREDICTED: protein phosphatase 2C 56-like [Jatropha curcas] |
| 2 |
Hb_002890_300 |
0.1155811382 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 3 |
Hb_002391_320 |
0.126283124 |
- |
- |
PREDICTED: phospholipase D delta-like [Jatropha curcas] |
| 4 |
Hb_005488_130 |
0.1385900045 |
- |
- |
transporter, putative [Ricinus communis] |
| 5 |
Hb_007101_260 |
0.1467297619 |
- |
- |
altered response to gravity (arg1), plant, putative [Ricinus communis] |
| 6 |
Hb_000753_110 |
0.1468791289 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor BIM1 isoform X6 [Jatropha curcas] |
| 7 |
Hb_000116_020 |
0.1476945292 |
- |
- |
PREDICTED: serine acetyltransferase 2-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000327_200 |
0.149132283 |
- |
- |
glutathione-s-transferase omega, putative [Ricinus communis] |
| 9 |
Hb_002641_120 |
0.1510847092 |
- |
- |
PREDICTED: calmodulin [Jatropha curcas] |
| 10 |
Hb_004052_100 |
0.1524381881 |
- |
- |
PREDICTED: protein SPIRAL1-like 3 [Jatropha curcas] |
| 11 |
Hb_006153_150 |
0.1526498772 |
- |
- |
PREDICTED: uncharacterized protein LOC105645999 [Jatropha curcas] |
| 12 |
Hb_008179_030 |
0.1528224604 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 8 [Jatropha curcas] |
| 13 |
Hb_000011_560 |
0.1553821616 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC2 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000032_510 |
0.1601628952 |
- |
- |
PREDICTED: SUN domain-containing protein 2 [Jatropha curcas] |
| 15 |
Hb_000069_360 |
0.1605710573 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_006153_160 |
0.1633116254 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 2A isoform X4 [Jatropha curcas] |
| 17 |
Hb_002016_140 |
0.1633830132 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase BAH1-like 1 [Jatropha curcas] |
| 18 |
Hb_001408_170 |
0.165828097 |
- |
- |
PREDICTED: phosphoglycolate phosphatase 2 [Jatropha curcas] |
| 19 |
Hb_000193_340 |
0.1673384369 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 isoform X1 [Jatropha curcas] |
| 20 |
Hb_091318_010 |
0.1695726246 |
- |
- |
hypothetical protein POPTR_0001s09520g [Populus trichocarpa] |