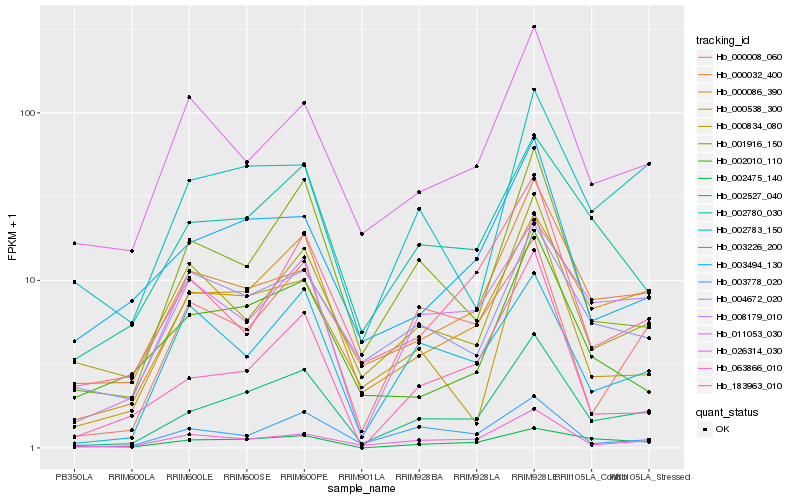
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002527_040 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat-containing protein 2, lrrc2, putative [Ricinus communis] |
| 2 |
Hb_000086_390 |
0.1211935276 |
- |
- |
PREDICTED: uncharacterized protein LOC105644503 [Jatropha curcas] |
| 3 |
Hb_002780_030 |
0.1522739179 |
- |
- |
PREDICTED: molybdate transporter 1 [Jatropha curcas] |
| 4 |
Hb_001916_150 |
0.153176499 |
transcription factor |
TF Family: NF-YB |
PREDICTED: nuclear transcription factor Y subunit B-3 [Vitis vinifera] |
| 5 |
Hb_026314_030 |
0.1587071931 |
- |
- |
PREDICTED: transmembrane protein 41B [Jatropha curcas] |
| 6 |
Hb_000032_400 |
0.1614434219 |
- |
- |
PREDICTED: UDP-glucose 4-epimerase GEPI48 [Jatropha curcas] |
| 7 |
Hb_008179_010 |
0.1689986843 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 16 [Jatropha curcas] |
| 8 |
Hb_003778_020 |
0.1718289504 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 9 |
Hb_003494_130 |
0.172546722 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 9-like [Jatropha curcas] |
| 10 |
Hb_063866_010 |
0.1755694702 |
- |
- |
PREDICTED: uncharacterized protein LOC105632605 [Jatropha curcas] |
| 11 |
Hb_002783_150 |
0.1767493309 |
- |
- |
cyclophilin [Hevea brasiliensis] |
| 12 |
Hb_000008_060 |
0.1780509537 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 13 |
Hb_000834_080 |
0.1803263981 |
- |
- |
PREDICTED: uncharacterized protein LOC105637718 [Jatropha curcas] |
| 14 |
Hb_002475_140 |
0.1815232344 |
- |
- |
PREDICTED: protein GAMETE EXPRESSED 1 [Jatropha curcas] |
| 15 |
Hb_002010_110 |
0.1821814648 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1 [Jatropha curcas] |
| 16 |
Hb_011053_030 |
0.1822371358 |
- |
- |
- |
| 17 |
Hb_000538_300 |
0.1854858081 |
- |
- |
PREDICTED: WAT1-related protein At3g02690, chloroplastic [Jatropha curcas] |
| 18 |
Hb_004672_020 |
0.1857121953 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 19 |
Hb_003226_200 |
0.1873439352 |
- |
- |
magnesium/proton exchanger, putative [Ricinus communis] |
| 20 |
Hb_183963_010 |
0.1882619687 |
- |
- |
core region of GTP cyclohydrolase I family protein [Populus trichocarpa] |