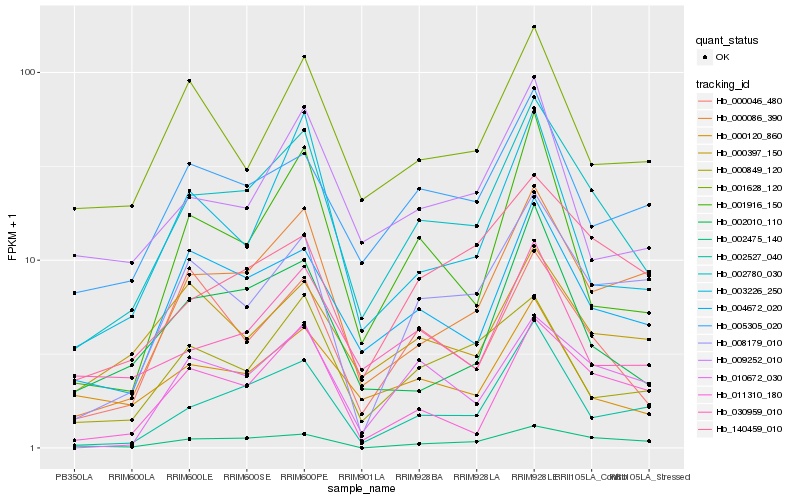
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002780_030 |
0.0 |
- |
- |
PREDICTED: molybdate transporter 1 [Jatropha curcas] |
| 2 |
Hb_004672_020 |
0.1313336035 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 3 |
Hb_000086_390 |
0.1401601734 |
- |
- |
PREDICTED: uncharacterized protein LOC105644503 [Jatropha curcas] |
| 4 |
Hb_005305_020 |
0.1482002389 |
- |
- |
PREDICTED: dicarboxylate transporter 1, chloroplastic [Jatropha curcas] |
| 5 |
Hb_002475_140 |
0.1512180564 |
- |
- |
PREDICTED: protein GAMETE EXPRESSED 1 [Jatropha curcas] |
| 6 |
Hb_002527_040 |
0.1522739179 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat-containing protein 2, lrrc2, putative [Ricinus communis] |
| 7 |
Hb_008179_010 |
0.1524990052 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 16 [Jatropha curcas] |
| 8 |
Hb_030959_010 |
0.1559081248 |
- |
- |
cullin, putative [Ricinus communis] |
| 9 |
Hb_000849_120 |
0.156190865 |
- |
- |
PREDICTED: linoleate 9S-lipoxygenase 6-like [Jatropha curcas] |
| 10 |
Hb_002010_110 |
0.1570893074 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1 [Jatropha curcas] |
| 11 |
Hb_001628_120 |
0.1583361495 |
- |
- |
putative ascorbate peroxidase, partial [Taraxacum brevicorniculatum] |
| 12 |
Hb_140459_010 |
0.1586089369 |
- |
- |
thioredoxin f-type, putative [Ricinus communis] |
| 13 |
Hb_000046_480 |
0.1592319609 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 5 [Jatropha curcas] |
| 14 |
Hb_000120_860 |
0.1600877436 |
- |
- |
nucellin, putative [Ricinus communis] |
| 15 |
Hb_001916_150 |
0.1612503028 |
transcription factor |
TF Family: NF-YB |
PREDICTED: nuclear transcription factor Y subunit B-3 [Vitis vinifera] |
| 16 |
Hb_009252_010 |
0.1623261351 |
- |
- |
PREDICTED: kinesin light chain [Jatropha curcas] |
| 17 |
Hb_011310_180 |
0.162352795 |
- |
- |
PREDICTED: phospholipase D alpha 1-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_003226_250 |
0.1651873936 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 2 [Jatropha curcas] |
| 19 |
Hb_010672_030 |
0.1663167019 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 11 [Jatropha curcas] |
| 20 |
Hb_000397_150 |
0.1664540335 |
- |
- |
PREDICTED: probable flavin-containing monooxygenase 1 [Jatropha curcas] |