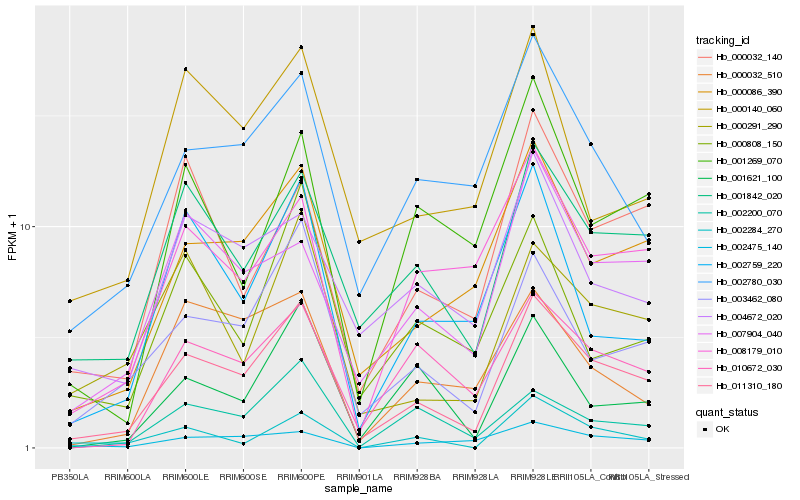
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011310_180 |
0.0 |
- |
- |
PREDICTED: phospholipase D alpha 1-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_003462_080 |
0.1338402778 |
- |
- |
Protein kinase APK1A, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_001842_020 |
0.1344093887 |
- |
- |
PREDICTED: low molecular weight phosphotyrosine protein phosphatase [Jatropha curcas] |
| 4 |
Hb_000086_390 |
0.1345716414 |
- |
- |
PREDICTED: uncharacterized protein LOC105644503 [Jatropha curcas] |
| 5 |
Hb_002780_030 |
0.162352795 |
- |
- |
PREDICTED: molybdate transporter 1 [Jatropha curcas] |
| 6 |
Hb_010672_030 |
0.1676231258 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 11 [Jatropha curcas] |
| 7 |
Hb_000291_290 |
0.1689580466 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_007904_040 |
0.1701214015 |
- |
- |
PREDICTED: uncharacterized protein LOC105644727 [Jatropha curcas] |
| 9 |
Hb_008179_010 |
0.1735604057 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 16 [Jatropha curcas] |
| 10 |
Hb_000032_140 |
0.17642883 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001621_100 |
0.1774130314 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase S.4 [Jatropha curcas] |
| 12 |
Hb_002200_070 |
0.1776001499 |
- |
- |
PREDICTED: endoglucanase 9-like [Populus euphratica] |
| 13 |
Hb_002759_220 |
0.1784612191 |
- |
- |
altered response to gravity (arg1), plant, putative [Ricinus communis] |
| 14 |
Hb_002475_140 |
0.1793954919 |
- |
- |
PREDICTED: protein GAMETE EXPRESSED 1 [Jatropha curcas] |
| 15 |
Hb_004672_020 |
0.179987422 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 16 |
Hb_000140_060 |
0.183305479 |
- |
- |
50S ribosomal protein L5, putative [Ricinus communis] |
| 17 |
Hb_000808_150 |
0.187455912 |
- |
- |
PREDICTED: uncharacterized protein LOC105647444 [Jatropha curcas] |
| 18 |
Hb_000032_510 |
0.1890563159 |
- |
- |
PREDICTED: SUN domain-containing protein 2 [Jatropha curcas] |
| 19 |
Hb_002284_270 |
0.1895377508 |
transcription factor |
TF Family: HB |
DNA binding protein, putative [Ricinus communis] |
| 20 |
Hb_001269_070 |
0.1895699354 |
- |
- |
double-stranded RNA binding protein, putative [Ricinus communis] |