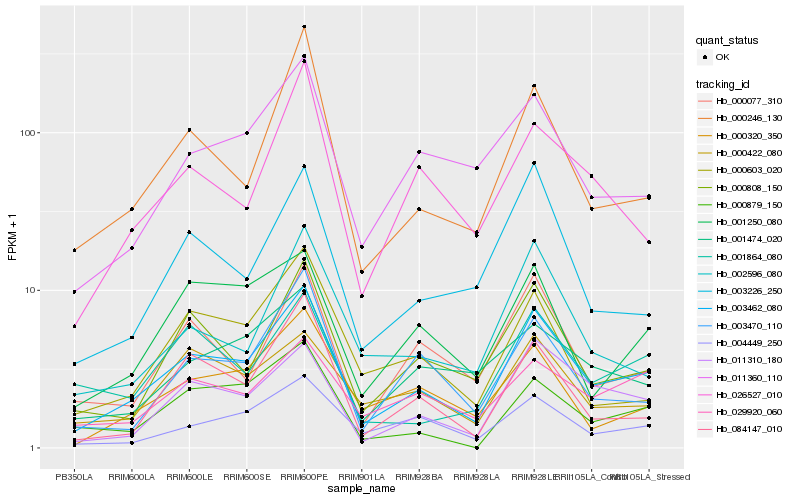
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003462_080 |
0.0 |
- |
- |
Protein kinase APK1A, chloroplast precursor, putative [Ricinus communis] |
| 2 |
Hb_011310_180 |
0.1338402778 |
- |
- |
PREDICTED: phospholipase D alpha 1-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_000808_150 |
0.1432504923 |
- |
- |
PREDICTED: uncharacterized protein LOC105647444 [Jatropha curcas] |
| 4 |
Hb_000246_130 |
0.1470201988 |
- |
- |
tubulin alpha chain, putative [Ricinus communis] |
| 5 |
Hb_004449_250 |
0.1502418079 |
- |
- |
PREDICTED: protein EI24 homolog [Jatropha curcas] |
| 6 |
Hb_011360_110 |
0.1517192438 |
- |
- |
PREDICTED: UDP-glucose 6-dehydrogenase 1 [Vitis vinifera] |
| 7 |
Hb_000320_350 |
0.1521150842 |
- |
- |
hypothetical protein RCOM_0904330 [Ricinus communis] |
| 8 |
Hb_000077_310 |
0.154967691 |
- |
- |
PREDICTED: clathrin light chain 1 [Jatropha curcas] |
| 9 |
Hb_084147_010 |
0.1574331823 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 10 |
Hb_003226_250 |
0.1575337829 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 2 [Jatropha curcas] |
| 11 |
Hb_003470_110 |
0.1583190636 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 12 |
Hb_001250_080 |
0.158370982 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g23390 [Jatropha curcas] |
| 13 |
Hb_002596_080 |
0.161977387 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At5g04160 [Jatropha curcas] |
| 14 |
Hb_026527_010 |
0.1630114769 |
- |
- |
PREDICTED: caffeoylshikimate esterase-like [Populus euphratica] |
| 15 |
Hb_000879_150 |
0.1631400415 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001474_020 |
0.1633940418 |
- |
- |
PREDICTED: uncharacterized protein LOC105650917 [Jatropha curcas] |
| 17 |
Hb_000422_080 |
0.1641952992 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor E2FB isoform X1 [Jatropha curcas] |
| 18 |
Hb_000603_020 |
0.1642154019 |
- |
- |
PREDICTED: uncharacterized protein LOC105638063 isoform X1 [Jatropha curcas] |
| 19 |
Hb_029920_060 |
0.1669132973 |
- |
- |
PREDICTED: UPF0554 protein-like isoform X2 [Jatropha curcas] |
| 20 |
Hb_001864_080 |
0.1670077871 |
- |
- |
PREDICTED: uncharacterized protein LOC105649405 [Jatropha curcas] |