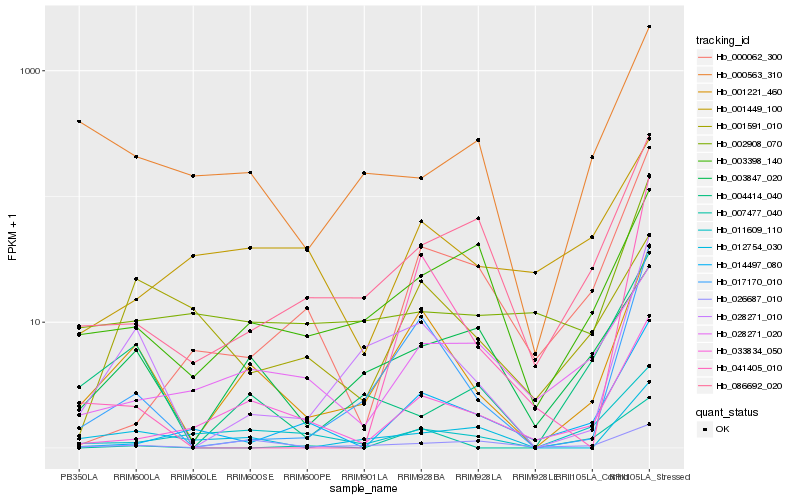
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001221_460 |
0.0 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 2 |
Hb_028271_010 |
0.182883032 |
- |
- |
PREDICTED: trans-resveratrol di-O-methyltransferase-like [Populus euphratica] |
| 3 |
Hb_017170_010 |
0.2076686035 |
- |
- |
PREDICTED: trans-resveratrol di-O-methyltransferase-like [Populus euphratica] |
| 4 |
Hb_033834_050 |
0.2556729156 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003847_020 |
0.2633144856 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 6 |
Hb_012754_030 |
0.2711562606 |
- |
- |
PREDICTED: uncharacterized protein LOC105633150 isoform X1 [Jatropha curcas] |
| 7 |
Hb_026687_010 |
0.2736124763 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Eucalyptus grandis] |
| 8 |
Hb_011609_110 |
0.2849523272 |
- |
- |
PREDICTED: T-complex protein 1 subunit gamma-like [Jatropha curcas] |
| 9 |
Hb_086692_020 |
0.2864511395 |
- |
- |
PREDICTED: protein kinase APK1B, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_003398_140 |
0.2870538721 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 11 |
Hb_004414_040 |
0.288148113 |
transcription factor |
TF Family: ERF |
AP2 domain-containing transcription factor family protein [Populus trichocarpa] |
| 12 |
Hb_002908_070 |
0.2997321748 |
- |
- |
DNA gyrase subunit A, chloroplast/mitochondrial precursor, putative [Ricinus communis] |
| 13 |
Hb_028271_020 |
0.3001040878 |
- |
- |
acyl-CoA oxidase, putative [Ricinus communis] |
| 14 |
Hb_000563_310 |
0.3078559217 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 3 [Hevea brasiliensis] |
| 15 |
Hb_007477_040 |
0.3087708646 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 16 |
Hb_014497_080 |
0.3106268486 |
- |
- |
PREDICTED: uncharacterized transporter YBR287W-like [Jatropha curcas] |
| 17 |
Hb_041405_010 |
0.3116268473 |
- |
- |
PREDICTED: trans-resveratrol di-O-methyltransferase-like [Glycine max] |
| 18 |
Hb_001591_010 |
0.3119171975 |
transcription factor |
TF Family: MYB |
myb domain protein [Hevea brasiliensis] |
| 19 |
Hb_000062_300 |
0.3155057719 |
- |
- |
PREDICTED: uncharacterized protein LOC105644761 [Jatropha curcas] |
| 20 |
Hb_001449_100 |
0.315568879 |
- |
- |
PREDICTED: transcription factor bHLH149 [Jatropha curcas] |