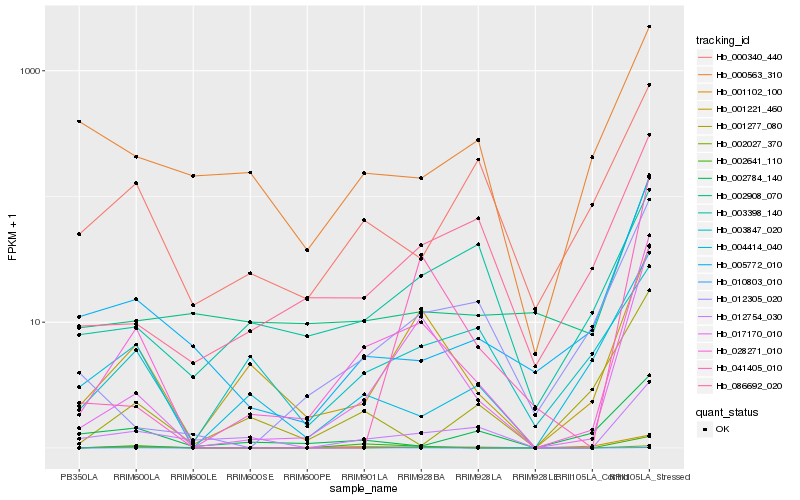
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_028271_010 |
0.0 |
- |
- |
PREDICTED: trans-resveratrol di-O-methyltransferase-like [Populus euphratica] |
| 2 |
Hb_001221_460 |
0.182883032 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 3 |
Hb_017170_010 |
0.1833102773 |
- |
- |
PREDICTED: trans-resveratrol di-O-methyltransferase-like [Populus euphratica] |
| 4 |
Hb_002641_110 |
0.2422510535 |
- |
- |
PREDICTED: uncharacterized protein LOC103448574 [Malus domestica] |
| 5 |
Hb_001102_100 |
0.2613153669 |
- |
- |
PREDICTED: uncharacterized protein LOC105140429 isoform X2 [Populus euphratica] |
| 6 |
Hb_012754_030 |
0.2637988287 |
- |
- |
PREDICTED: uncharacterized protein LOC105633150 isoform X1 [Jatropha curcas] |
| 7 |
Hb_004414_040 |
0.2763264066 |
transcription factor |
TF Family: ERF |
AP2 domain-containing transcription factor family protein [Populus trichocarpa] |
| 8 |
Hb_005772_010 |
0.2922667312 |
- |
- |
PREDICTED: abscisic acid receptor PYL4 [Jatropha curcas] |
| 9 |
Hb_086692_020 |
0.2950611677 |
- |
- |
PREDICTED: protein kinase APK1B, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_003847_020 |
0.3037884698 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 11 |
Hb_000340_440 |
0.3048857826 |
- |
- |
PREDICTED: gibberellin receptor GID1B [Jatropha curcas] |
| 12 |
Hb_010803_010 |
0.3096448895 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 13 |
Hb_002908_070 |
0.3106927628 |
- |
- |
DNA gyrase subunit A, chloroplast/mitochondrial precursor, putative [Ricinus communis] |
| 14 |
Hb_012305_020 |
0.3134207721 |
- |
- |
beta-1,3-glucanase [Hevea brasiliensis] |
| 15 |
Hb_041405_010 |
0.315512664 |
- |
- |
PREDICTED: trans-resveratrol di-O-methyltransferase-like [Glycine max] |
| 16 |
Hb_002784_140 |
0.316974479 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 17 |
Hb_003398_140 |
0.317589602 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_001277_080 |
0.3202340468 |
- |
- |
PREDICTED: vegetative cell wall protein gp1 [Jatropha curcas] |
| 19 |
Hb_000563_310 |
0.3240466775 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 3 [Hevea brasiliensis] |
| 20 |
Hb_002027_370 |
0.3354114347 |
- |
- |
conserved hypothetical protein [Ricinus communis] |