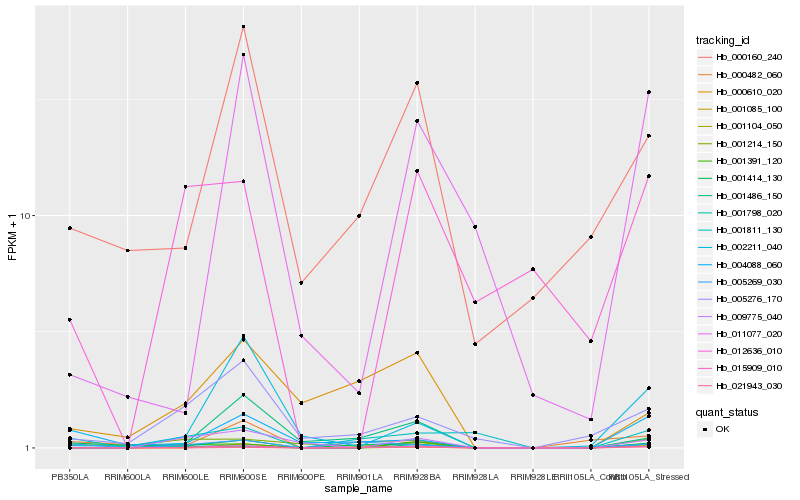
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_015909_010 |
0.0 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 2 |
Hb_005269_030 |
0.271350321 |
- |
- |
cysteine protease, putative [Ricinus communis] |
| 3 |
Hb_001391_120 |
0.2896053607 |
- |
- |
Acetyl esterase, putative [Ricinus communis] |
| 4 |
Hb_001414_130 |
0.2947512148 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 5 |
Hb_001798_020 |
0.3041341838 |
- |
- |
PREDICTED: uncharacterized protein LOC104594032 [Nelumbo nucifera] |
| 6 |
Hb_000160_240 |
0.3088944285 |
- |
- |
PREDICTED: uncharacterized protein At1g66480 [Jatropha curcas] |
| 7 |
Hb_005276_170 |
0.3104845542 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 2 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001811_130 |
0.3108049441 |
- |
- |
- |
| 9 |
Hb_004088_060 |
0.312549103 |
- |
- |
hypothetical protein POPTR_0012s01875g [Populus trichocarpa] |
| 10 |
Hb_001214_150 |
0.3210337872 |
- |
- |
PREDICTED: uncharacterized protein LOC102624288 [Citrus sinensis] |
| 11 |
Hb_001104_050 |
0.3213749518 |
- |
- |
- |
| 12 |
Hb_000610_020 |
0.328126324 |
- |
- |
unknown [Glycine max] |
| 13 |
Hb_000482_060 |
0.3447944025 |
transcription factor |
TF Family: LOB |
hypothetical protein JCGZ_07664 [Jatropha curcas] |
| 14 |
Hb_001486_150 |
0.3459145253 |
- |
- |
- |
| 15 |
Hb_021943_030 |
0.3513594186 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 16 |
Hb_012636_010 |
0.3539773576 |
- |
- |
- |
| 17 |
Hb_009775_040 |
0.3603348643 |
- |
- |
PREDICTED: probable glutathione S-transferase parC [Vitis vinifera] |
| 18 |
Hb_011077_020 |
0.3638125353 |
- |
- |
kinase, putative [Ricinus communis] |
| 19 |
Hb_001085_100 |
0.3674677454 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 20 |
Hb_002211_040 |
0.3677440897 |
- |
- |
PREDICTED: subtilisin-like protease SBT3.5 [Jatropha curcas] |