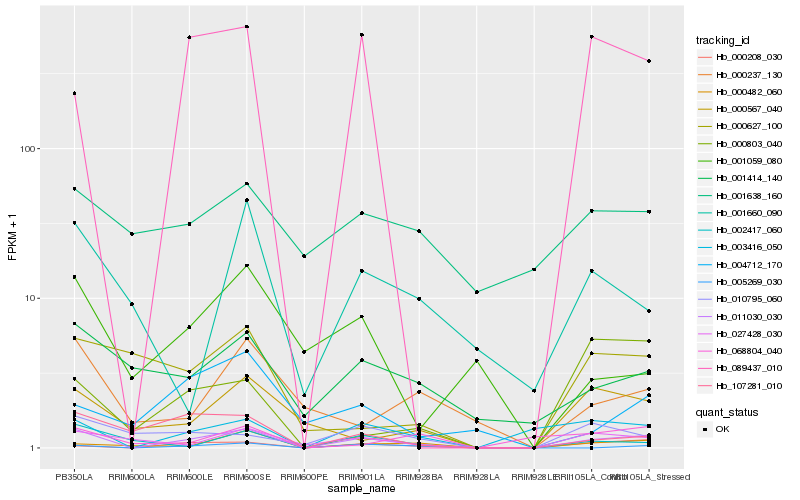
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011030_030 |
0.0 |
- |
- |
PLC-like phosphodiesterases superfamily protein [Theobroma cacao] |
| 2 |
Hb_000208_030 |
0.2527424014 |
- |
- |
- |
| 3 |
Hb_003416_050 |
0.2693539527 |
- |
- |
- |
| 4 |
Hb_089437_010 |
0.2739988071 |
- |
- |
PREDICTED: polyadenylate-binding protein-interacting protein 10-like isoform X3 [Jatropha curcas] |
| 5 |
Hb_005269_030 |
0.2903107977 |
- |
- |
cysteine protease, putative [Ricinus communis] |
| 6 |
Hb_027428_030 |
0.295854587 |
- |
- |
unnamed protein product [Coffea canephora] |
| 7 |
Hb_000482_060 |
0.304286763 |
transcription factor |
TF Family: LOB |
hypothetical protein JCGZ_07664 [Jatropha curcas] |
| 8 |
Hb_107281_010 |
0.3071475019 |
- |
- |
protein with unknown function [Ricinus communis] |
| 9 |
Hb_002417_060 |
0.3095231842 |
- |
- |
Cell division cycle protein, putative [Ricinus communis] |
| 10 |
Hb_001414_140 |
0.3162300577 |
- |
- |
PREDICTED: probable S-adenosylmethionine-dependent methyltransferase At5g38100 [Populus euphratica] |
| 11 |
Hb_000567_040 |
0.324165859 |
- |
- |
PREDICTED: uncharacterized protein LOC105649657 [Jatropha curcas] |
| 12 |
Hb_010795_060 |
0.3352038881 |
- |
- |
- |
| 13 |
Hb_000237_130 |
0.3407743103 |
- |
- |
- |
| 14 |
Hb_000627_100 |
0.3439421293 |
- |
- |
- |
| 15 |
Hb_004712_170 |
0.3443790015 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF181 [Jatropha curcas] |
| 16 |
Hb_001660_090 |
0.3456383959 |
- |
- |
PREDICTED: stromal 70 kDa heat shock-related protein, chloroplastic-like [Jatropha curcas] |
| 17 |
Hb_001059_080 |
0.3509219457 |
- |
- |
PREDICTED: F-box protein At1g61340-like [Musa acuminata subsp. malaccensis] |
| 18 |
Hb_000803_040 |
0.3512019597 |
- |
- |
- |
| 19 |
Hb_068804_040 |
0.3516699498 |
- |
- |
- |
| 20 |
Hb_001638_160 |
0.3517123402 |
- |
- |
PREDICTED: uncharacterized protein LOC105642785 [Jatropha curcas] |