| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
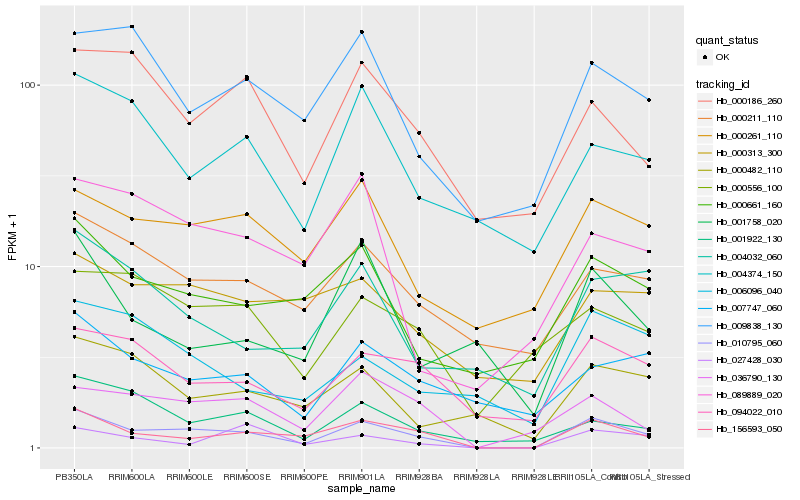
Hb_010795_060 |
0.0 |
- |
- |
- |
| 2 |
Hb_156593_050 |
0.139555564 |
- |
- |
PREDICTED: DNA-binding protein RHL1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_036790_130 |
0.2065179986 |
- |
- |
- |
| 4 |
Hb_094022_010 |
0.2066299627 |
transcription factor |
TF Family: C2C2-GATA |
hypothetical protein JCGZ_05054 [Jatropha curcas] |
| 5 |
Hb_000661_160 |
0.2108282059 |
- |
- |
hexokinase [Manihot esculenta] |
| 6 |
Hb_000261_110 |
0.2151910466 |
- |
- |
PREDICTED: probable methionine--tRNA ligase [Jatropha curcas] |
| 7 |
Hb_000211_110 |
0.2173193513 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 35 kDa protein isoform X1 [Jatropha curcas] |
| 8 |
Hb_009838_130 |
0.2215359059 |
- |
- |
hypothetical protein CISIN_1g010403mg [Citrus sinensis] |
| 9 |
Hb_001922_130 |
0.22290133 |
- |
- |
hypothetical protein PHAVU_002G294300g [Phaseolus vulgaris] |
| 10 |
Hb_000482_110 |
0.2231205264 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 11 |
Hb_004032_060 |
0.2250030888 |
- |
- |
elongation factor 1 gamma-like protein, partial [Ipomoea nil] |
| 12 |
Hb_007747_060 |
0.2256631351 |
- |
- |
PREDICTED: mitochondrial fission protein ELM1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_004374_150 |
0.2256944618 |
- |
- |
PREDICTED: uncharacterized protein LOC105650290 isoform X1 [Jatropha curcas] |
| 14 |
Hb_027428_030 |
0.227048811 |
- |
- |
unnamed protein product [Coffea canephora] |
| 15 |
Hb_000186_260 |
0.2281055308 |
transcription factor |
TF Family: PHD |
PREDICTED: protein polybromo-1-like [Jatropha curcas] |
| 16 |
Hb_000556_100 |
0.2286149014 |
- |
- |
PREDICTED: uncharacterized protein LOC104784782 isoform X2 [Camelina sativa] |
| 17 |
Hb_006096_040 |
0.230351001 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 8 [Jatropha curcas] |
| 18 |
Hb_089889_020 |
0.2307220149 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein At2g36080-like [Jatropha curcas] |
| 19 |
Hb_001758_020 |
0.2310965989 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000313_300 |
0.2341747361 |
- |
- |
PREDICTED: F-box protein SKIP19-like [Jatropha curcas] |