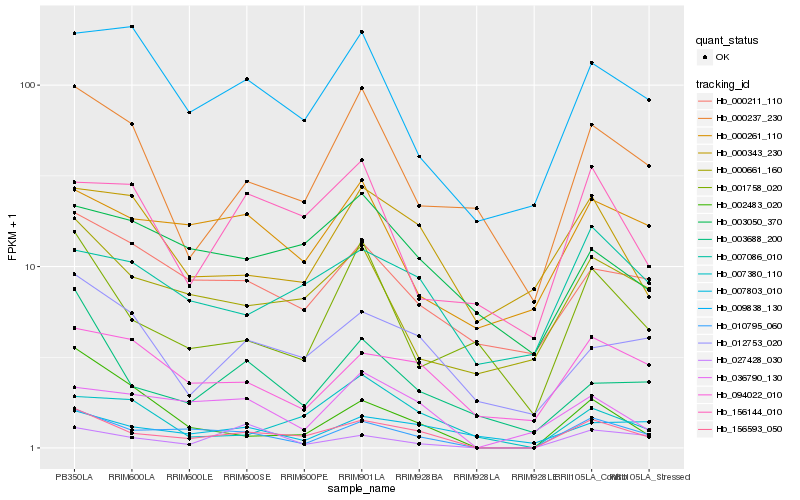
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_156593_050 |
0.0 |
- |
- |
PREDICTED: DNA-binding protein RHL1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_010795_060 |
0.139555564 |
- |
- |
- |
| 3 |
Hb_012753_020 |
0.20916386 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000661_160 |
0.2134453411 |
- |
- |
hexokinase [Manihot esculenta] |
| 5 |
Hb_094022_010 |
0.2159664332 |
transcription factor |
TF Family: C2C2-GATA |
hypothetical protein JCGZ_05054 [Jatropha curcas] |
| 6 |
Hb_036790_130 |
0.2159948512 |
- |
- |
- |
| 7 |
Hb_007086_010 |
0.2190860485 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 26-like [Jatropha curcas] |
| 8 |
Hb_003688_200 |
0.2202347371 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000237_230 |
0.2235506845 |
- |
- |
PREDICTED: RNA polymerase I-specific transcription initiation factor RRN3 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000211_110 |
0.2255367026 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 35 kDa protein isoform X1 [Jatropha curcas] |
| 11 |
Hb_001758_020 |
0.2258900489 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_009838_130 |
0.2294179136 |
- |
- |
hypothetical protein CISIN_1g010403mg [Citrus sinensis] |
| 13 |
Hb_027428_030 |
0.2304172174 |
- |
- |
unnamed protein product [Coffea canephora] |
| 14 |
Hb_000343_230 |
0.232713461 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase PASTICCINO1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002483_020 |
0.2335407293 |
- |
- |
- |
| 16 |
Hb_007803_010 |
0.2339370846 |
- |
- |
PREDICTED: F-box protein At3g07870-like [Jatropha curcas] |
| 17 |
Hb_007380_110 |
0.2343881296 |
- |
- |
hypothetical protein JCGZ_16415 [Jatropha curcas] |
| 18 |
Hb_003050_370 |
0.2354475539 |
- |
- |
mitochondrial inner membrane protease subunit, putative [Ricinus communis] |
| 19 |
Hb_000261_110 |
0.2361474473 |
- |
- |
PREDICTED: probable methionine--tRNA ligase [Jatropha curcas] |
| 20 |
Hb_156144_010 |
0.2372385197 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA9 [Jatropha curcas] |