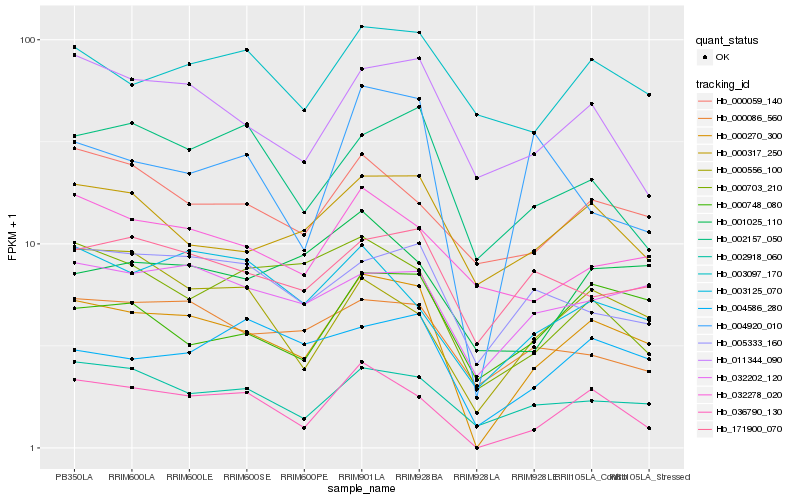
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000270_300 |
0.0 |
- |
- |
- |
| 2 |
Hb_036790_130 |
0.1259033412 |
- |
- |
- |
| 3 |
Hb_002918_060 |
0.1562082126 |
- |
- |
PREDICTED: glyoxysomal processing protease, glyoxysomal isoform X1 [Jatropha curcas] |
| 4 |
Hb_032202_120 |
0.1607105382 |
- |
- |
PREDICTED: uncharacterized protein LOC105643059 isoform X2 [Jatropha curcas] |
| 5 |
Hb_004586_280 |
0.1625838279 |
- |
- |
PREDICTED: GEM-like protein 1 [Jatropha curcas] |
| 6 |
Hb_171900_070 |
0.1681222739 |
- |
- |
- |
| 7 |
Hb_000317_250 |
0.1686420123 |
- |
- |
PREDICTED: calcium-dependent protein kinase SK5 isoform X1 [Jatropha curcas] |
| 8 |
Hb_011344_090 |
0.1686424943 |
- |
- |
PREDICTED: FAM10 family protein At4g22670 [Jatropha curcas] |
| 9 |
Hb_000748_080 |
0.1709882365 |
- |
- |
hypothetical protein OsI_28173 [Oryza sativa Indica Group] |
| 10 |
Hb_005333_160 |
0.1722091907 |
- |
- |
hypothetical protein B456_009G230200 [Gossypium raimondii] |
| 11 |
Hb_004920_010 |
0.1764473692 |
- |
- |
PREDICTED: universal stress protein A-like protein [Jatropha curcas] |
| 12 |
Hb_003125_070 |
0.1765253387 |
- |
- |
PREDICTED: serine/threonine-protein kinase rio1-like [Jatropha curcas] |
| 13 |
Hb_000703_210 |
0.1766346831 |
- |
- |
Inactive protein kinase [Gossypium arboreum] |
| 14 |
Hb_000086_560 |
0.1766557704 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase SUVR2 isoform X2 [Jatropha curcas] |
| 15 |
Hb_002157_050 |
0.1768810872 |
- |
- |
hypothetical protein JCGZ_16907 [Jatropha curcas] |
| 16 |
Hb_000556_100 |
0.177918445 |
- |
- |
PREDICTED: uncharacterized protein LOC104784782 isoform X2 [Camelina sativa] |
| 17 |
Hb_032278_020 |
0.1782542502 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 16 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000059_140 |
0.178501928 |
- |
- |
rnase h, putative [Ricinus communis] |
| 19 |
Hb_001025_110 |
0.17978818 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003097_170 |
0.1803630099 |
- |
- |
PREDICTED: protein EARLY RESPONSIVE TO DEHYDRATION 15-like isoform X3 [Jatropha curcas] |