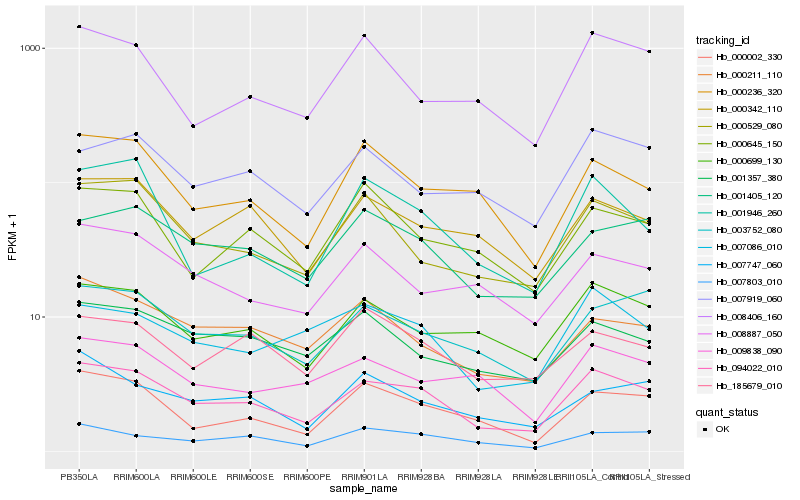
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_094022_010 |
0.0 |
transcription factor |
TF Family: C2C2-GATA |
hypothetical protein JCGZ_05054 [Jatropha curcas] |
| 2 |
Hb_000699_130 |
0.1088553183 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003752_080 |
0.1122194605 |
- |
- |
PREDICTED: ubiquitin-like-conjugating enzyme ATG10 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000529_080 |
0.113757753 |
- |
- |
PREDICTED: uncharacterized protein LOC105641675 [Jatropha curcas] |
| 5 |
Hb_000002_330 |
0.114465704 |
- |
- |
PREDICTED: actin-related protein 9 [Jatropha curcas] |
| 6 |
Hb_000342_110 |
0.1171290781 |
- |
- |
PREDICTED: uncharacterized protein LOC105638174 [Jatropha curcas] |
| 7 |
Hb_007803_010 |
0.1183950355 |
- |
- |
PREDICTED: F-box protein At3g07870-like [Jatropha curcas] |
| 8 |
Hb_001405_120 |
0.1191494353 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit H [Jatropha curcas] |
| 9 |
Hb_000211_110 |
0.1208270978 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 35 kDa protein isoform X1 [Jatropha curcas] |
| 10 |
Hb_007086_010 |
0.1241496466 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 26-like [Jatropha curcas] |
| 11 |
Hb_001357_380 |
0.1246266067 |
- |
- |
PREDICTED: probable tetraacyldisaccharide 4'-kinase, mitochondrial isoform X1 [Jatropha curcas] |
| 12 |
Hb_000236_320 |
0.1268887201 |
- |
- |
PREDICTED: ER membrane protein complex subunit 8/9 homolog [Jatropha curcas] |
| 13 |
Hb_008406_160 |
0.1290003801 |
- |
- |
PREDICTED: ADP,ATP carrier protein 3, mitochondrial [Jatropha curcas] |
| 14 |
Hb_008887_050 |
0.1299130557 |
- |
- |
PREDICTED: DNA-binding protein BIN4 [Jatropha curcas] |
| 15 |
Hb_185679_010 |
0.1306251009 |
- |
- |
serine/threonine-protein kinase rio2, putative [Ricinus communis] |
| 16 |
Hb_007919_060 |
0.1313785482 |
- |
- |
PREDICTED: UPF0047 protein C4A8.02c [Jatropha curcas] |
| 17 |
Hb_001946_260 |
0.1320528625 |
- |
- |
PREDICTED: ribosome biogenesis regulatory protein homolog isoform X1 [Jatropha curcas] |
| 18 |
Hb_007747_060 |
0.1321687175 |
- |
- |
PREDICTED: mitochondrial fission protein ELM1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_009838_090 |
0.1321949636 |
- |
- |
PREDICTED: uncharacterized protein LOC105638421 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000645_150 |
0.1321949896 |
- |
- |
zinc finger protein, putative [Ricinus communis] |